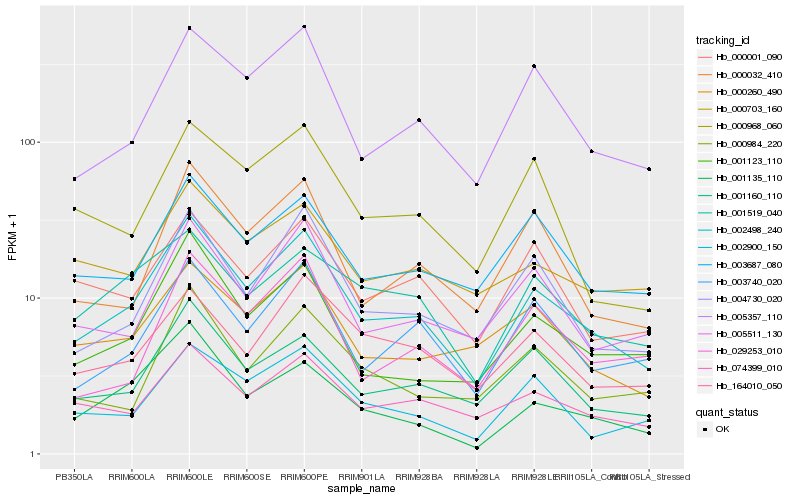
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002498_240 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005357_110 |
0.1084254245 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 3 |
Hb_001160_110 |
0.111077018 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 4 |
Hb_004730_020 |
0.1122034323 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 5 |
Hb_074399_010 |
0.1130320139 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 6 |
Hb_001123_110 |
0.1147320817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003740_020 |
0.1187622627 |
- |
- |
PREDICTED: methylglutaconyl-CoA hydratase, mitochondrial isoform X2 [Jatropha curcas] |
| 8 |
Hb_005511_130 |
0.1193014989 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 9 |
Hb_000032_410 |
0.1230410226 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 10 |
Hb_002900_150 |
0.1230634653 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |
| 11 |
Hb_001135_110 |
0.1243802271 |
- |
- |
- |
| 12 |
Hb_000260_490 |
0.1252572159 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 13 |
Hb_003687_080 |
0.1254015765 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 14 |
Hb_164010_050 |
0.1279655761 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 15 |
Hb_001519_040 |
0.1327315169 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 16 |
Hb_000001_090 |
0.1327740125 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 17 |
Hb_000984_220 |
0.1350117944 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 18 |
Hb_029253_010 |
0.1350666334 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 19 |
Hb_000703_160 |
0.1351210279 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000968_060 |
0.135396976 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |