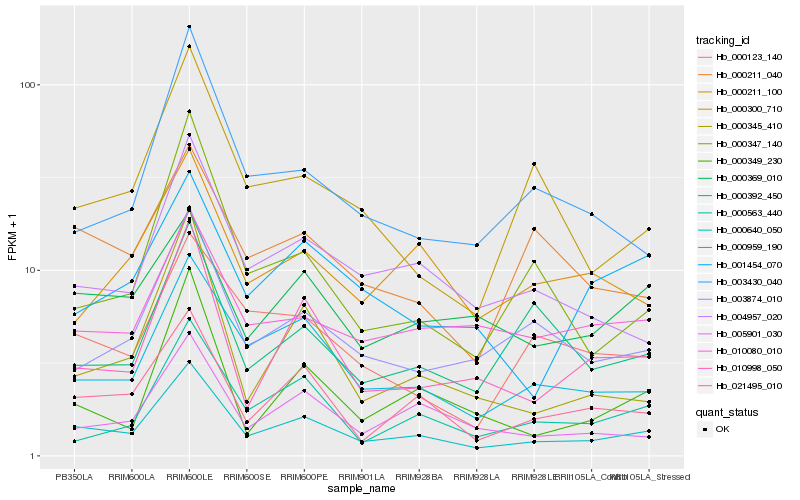
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000640_050 |
0.0 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 2 |
Hb_021495_010 |
0.1140942403 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000959_190 |
0.1191647324 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 4 |
Hb_010998_050 |
0.1377983707 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004957_020 |
0.1414578557 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005901_030 |
0.1439622875 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003874_010 |
0.1466167034 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_010080_010 |
0.1554359548 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 9 |
Hb_000369_010 |
0.1573488069 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000563_440 |
0.1573599814 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000123_140 |
0.162187021 |
- |
- |
PREDICTED: 21 kDa protein-like [Populus euphratica] |
| 12 |
Hb_000345_410 |
0.162580082 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 13 |
Hb_003430_040 |
0.1642074927 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 14 |
Hb_000211_040 |
0.1645511219 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 15 |
Hb_000347_140 |
0.1649531168 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_000349_230 |
0.1651774391 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 17 |
Hb_000300_710 |
0.1655433495 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 18 |
Hb_000392_450 |
0.1658089834 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000211_100 |
0.1667468416 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 20 |
Hb_001454_070 |
0.1698527231 |
- |
- |
Uveal autoantigen with coiled-coil domains and ankyrin repeats isoform 1 [Theobroma cacao] |