| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
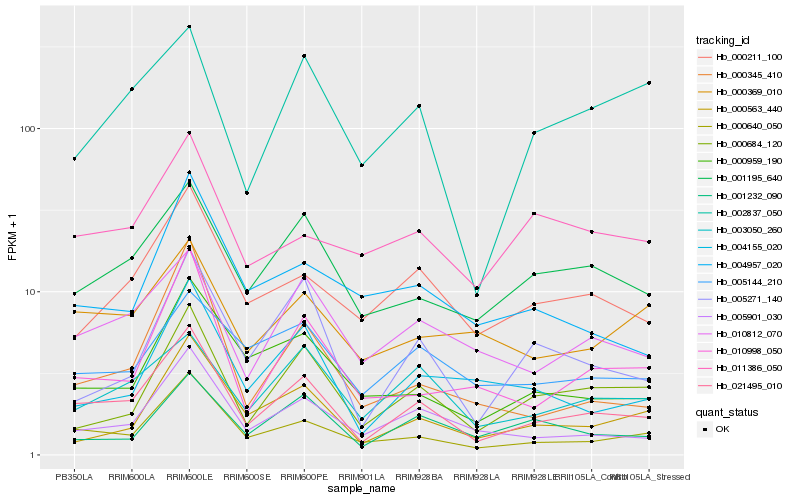
Hb_021495_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000640_050 |
0.1140942403 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 3 |
Hb_005901_030 |
0.1412135042 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001195_640 |
0.1499041807 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 5 |
Hb_010812_070 |
0.1510548377 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein NETWORKED 4A [Jatropha curcas] |
| 6 |
Hb_000211_100 |
0.1586734871 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 7 |
Hb_003050_260 |
0.1602315059 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 8 |
Hb_000684_120 |
0.1616781027 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000959_190 |
0.162430802 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 10 |
Hb_010998_050 |
0.1659939003 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000563_440 |
0.1691954867 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_005271_140 |
0.1726610888 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3 [Morus notabilis] |
| 13 |
Hb_004155_020 |
0.1728289694 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 14 |
Hb_005144_210 |
0.1786435894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000369_010 |
0.1787039371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001232_090 |
0.1789181074 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 17 |
Hb_000345_410 |
0.180568848 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 18 |
Hb_002837_050 |
0.1812658999 |
- |
- |
hypothetical protein POPTR_0006s00550g [Populus trichocarpa] |
| 19 |
Hb_011386_050 |
0.184473744 |
- |
- |
PREDICTED: malonyl-CoA-acyl carrier protein transacylase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_004957_020 |
0.1851658615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |