| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
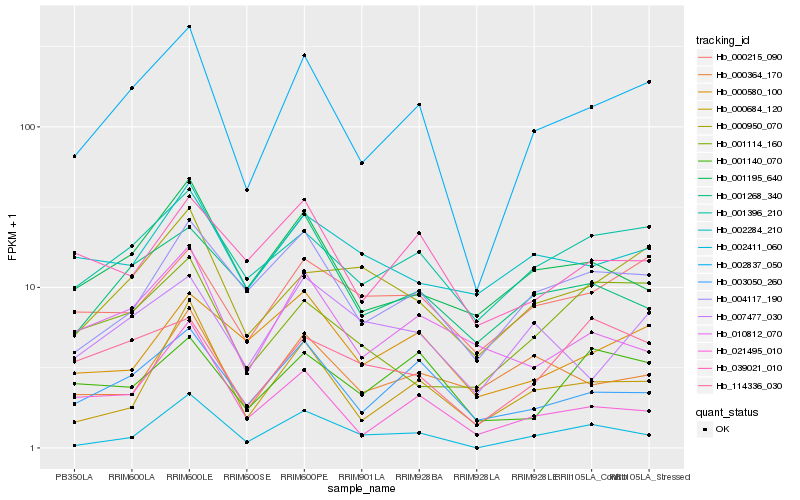
Hb_002837_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s00550g [Populus trichocarpa] |
| 2 |
Hb_001396_210 |
0.1402992673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003050_260 |
0.1423886919 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 4 |
Hb_004117_190 |
0.1595735013 |
- |
- |
PREDICTED: uncharacterized protein LOC105649109 [Jatropha curcas] |
| 5 |
Hb_000684_120 |
0.1654744449 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001195_640 |
0.1667392612 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 7 |
Hb_000950_070 |
0.1699104988 |
- |
- |
PREDICTED: uncharacterized protein LOC105645259 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000215_090 |
0.1738950569 |
- |
- |
cytochrome b5 domain-containing family protein [Populus trichocarpa] |
| 9 |
Hb_007477_030 |
0.1747212052 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 10 |
Hb_021495_010 |
0.1812658999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001268_340 |
0.1827944094 |
- |
- |
Actin, putative [Ricinus communis] |
| 12 |
Hb_114336_030 |
0.1843624575 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 13 |
Hb_010812_070 |
0.1849956342 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein NETWORKED 4A [Jatropha curcas] |
| 14 |
Hb_000580_100 |
0.1870288706 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002411_060 |
0.1880040339 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 16 |
Hb_001140_070 |
0.1901464096 |
- |
- |
Chromosome transmission fidelity protein 8, putative isoform 2 [Theobroma cacao] |
| 17 |
Hb_039021_010 |
0.1908934749 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 18 |
Hb_002284_210 |
0.1909136888 |
- |
- |
PREDICTED: protein canopy-1 [Jatropha curcas] |
| 19 |
Hb_001114_160 |
0.1931036275 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000364_170 |
0.1933518642 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |