| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
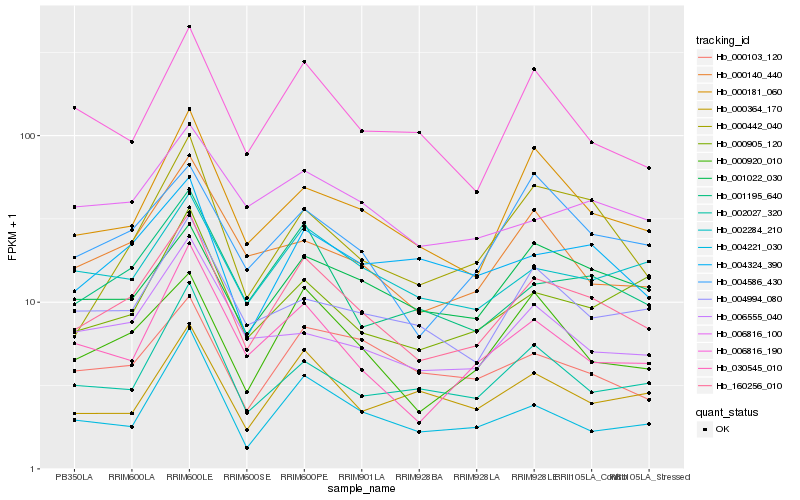
Hb_160256_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001195_640 |
0.1012484806 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 3 |
Hb_000920_010 |
0.1087150071 |
- |
- |
hypothetical protein PRUPE_ppa013940mg [Prunus persica] |
| 4 |
Hb_000181_060 |
0.1158991216 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002284_210 |
0.1196250756 |
- |
- |
PREDICTED: protein canopy-1 [Jatropha curcas] |
| 6 |
Hb_030545_010 |
0.1198803714 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 7 |
Hb_000140_440 |
0.1248436572 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 8 |
Hb_004324_390 |
0.1265096687 |
- |
- |
PREDICTED: plastidial pyruvate kinase 2 [Jatropha curcas] |
| 9 |
Hb_004221_030 |
0.1273801402 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000905_120 |
0.1282748876 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 11 |
Hb_002027_320 |
0.1285957573 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 12 |
Hb_006816_190 |
0.1293816601 |
- |
- |
calreticulin family protein [Populus trichocarpa] |
| 13 |
Hb_004586_430 |
0.1294123755 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 14 |
Hb_006816_100 |
0.1294842117 |
- |
- |
hypothetical protein CISIN_1g0373181mg, partial [Citrus sinensis] |
| 15 |
Hb_004994_080 |
0.1304542146 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_001022_030 |
0.1320400692 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 17 |
Hb_000103_120 |
0.1323105102 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000442_040 |
0.1323228429 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000364_170 |
0.1358987315 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 20 |
Hb_006555_040 |
0.1375253572 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |