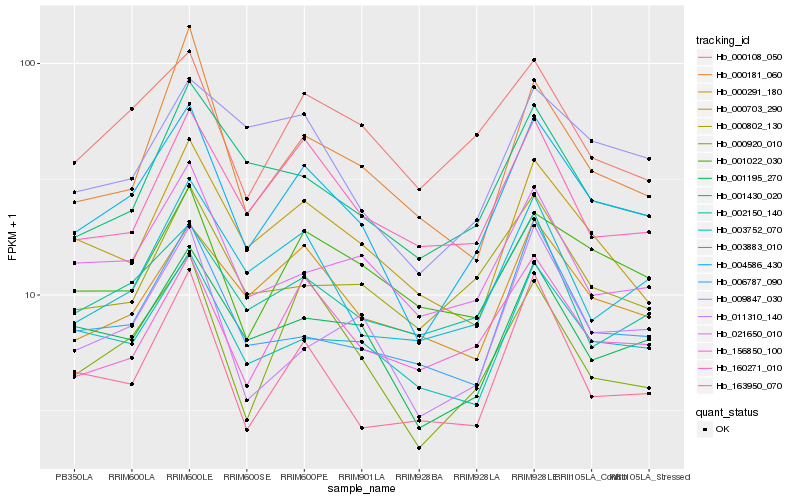
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004586_430 |
0.0 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001195_270 |
0.0957585888 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000920_010 |
0.1036831577 |
- |
- |
hypothetical protein PRUPE_ppa013940mg [Prunus persica] |
| 4 |
Hb_003752_070 |
0.1049648187 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 5 |
Hb_156850_100 |
0.1117687097 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 6 |
Hb_011310_140 |
0.1118879773 |
- |
- |
hypothetical protein JCGZ_07173 [Jatropha curcas] |
| 7 |
Hb_003883_010 |
0.1127352878 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 8 |
Hb_021650_010 |
0.1156531228 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 9 |
Hb_000108_050 |
0.1159941578 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 10 |
Hb_000703_290 |
0.1163407499 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 11 |
Hb_000291_180 |
0.1163741708 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 12 |
Hb_001022_030 |
0.116884993 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 13 |
Hb_160271_010 |
0.1173455573 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_006787_090 |
0.1193563785 |
- |
- |
PREDICTED: uncharacterized protein LOC105646282 [Jatropha curcas] |
| 15 |
Hb_001430_020 |
0.1195303784 |
- |
- |
Actin-like ATPase superfamily protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_009847_030 |
0.1196185698 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 17 |
Hb_163950_070 |
0.1199171932 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 18 |
Hb_000802_130 |
0.1222186753 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 19 |
Hb_002150_140 |
0.1231221993 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 20 |
Hb_000181_060 |
0.1232171217 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |