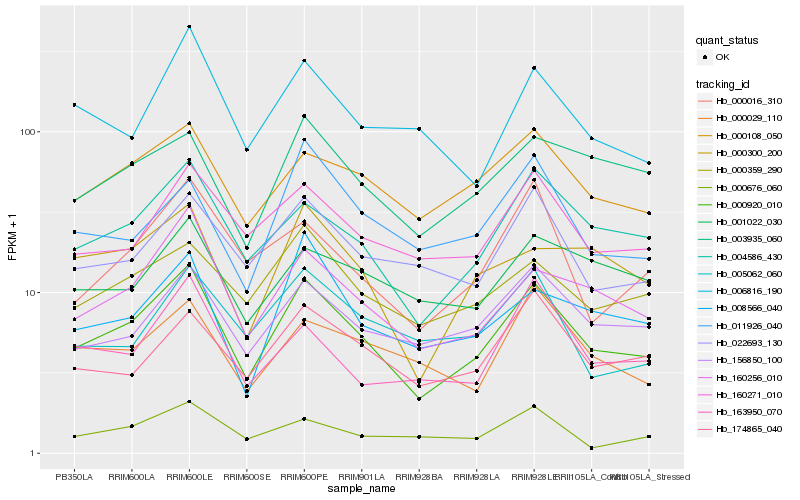
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000920_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa013940mg [Prunus persica] |
| 2 |
Hb_004586_430 |
0.1036831577 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 3 |
Hb_160256_010 |
0.1087150071 |
- |
- |
- |
| 4 |
Hb_000108_050 |
0.1216134166 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 5 |
Hb_156850_100 |
0.1231090295 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 6 |
Hb_160271_010 |
0.1246864326 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_022693_130 |
0.1292674201 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000359_290 |
0.1321882883 |
- |
- |
Triose phosphate/phosphate translocator, non-green plastid, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_174865_040 |
0.1322265245 |
- |
- |
PREDICTED: crt homolog 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000300_200 |
0.1339074086 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 11 |
Hb_003935_060 |
0.1340592803 |
- |
- |
PREDICTED: very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase PASTICCINO 2 [Jatropha curcas] |
| 12 |
Hb_008566_040 |
0.1389881851 |
- |
- |
PREDICTED: uncharacterized protein LOC105647590 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000016_310 |
0.1408242404 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_006816_190 |
0.1411725737 |
- |
- |
calreticulin family protein [Populus trichocarpa] |
| 15 |
Hb_000676_060 |
0.1423406954 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 16 |
Hb_000029_110 |
0.1445038816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_163950_070 |
0.1448625217 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 18 |
Hb_005062_060 |
0.1450961281 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 19 |
Hb_001022_030 |
0.1465273915 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 20 |
Hb_011926_040 |
0.1476186908 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |