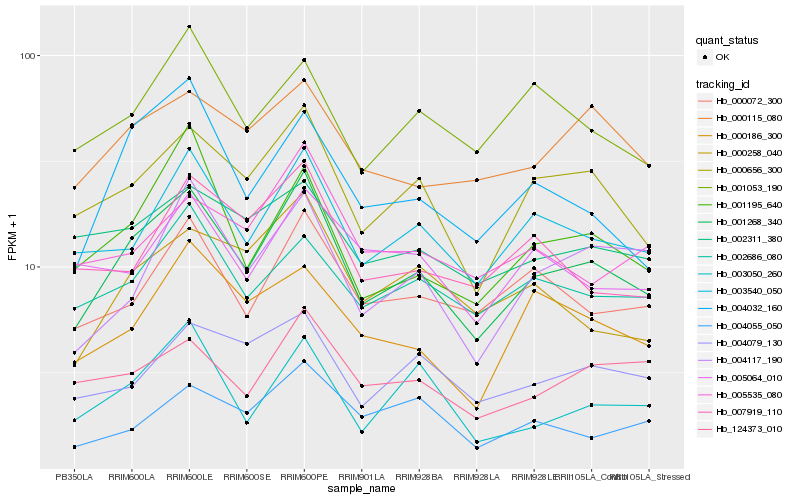
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001268_340 |
0.0 |
- |
- |
Actin, putative [Ricinus communis] |
| 2 |
Hb_000656_300 |
0.0957587516 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 7 member B4-like [Populus euphratica] |
| 3 |
Hb_003540_050 |
0.1048012288 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 4 |
Hb_001195_640 |
0.1208113681 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 5 |
Hb_004032_160 |
0.1214494259 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 6 |
Hb_000072_300 |
0.1227553901 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 7 |
Hb_004117_190 |
0.1247383813 |
- |
- |
PREDICTED: uncharacterized protein LOC105649109 [Jatropha curcas] |
| 8 |
Hb_000258_040 |
0.1286905714 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 9 |
Hb_000115_080 |
0.1302479278 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |
| 10 |
Hb_002686_080 |
0.1308447238 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein ING2-like [Jatropha curcas] |
| 11 |
Hb_004079_130 |
0.1319564951 |
- |
- |
PREDICTED: uncharacterized protein LOC105637708 [Jatropha curcas] |
| 12 |
Hb_124373_010 |
0.1333750377 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X1 [Jatropha curcas] |
| 13 |
Hb_000186_300 |
0.1344617066 |
- |
- |
PREDICTED: malonyl-CoA decarboxylase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_003050_260 |
0.1348864789 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 15 |
Hb_007919_110 |
0.135858288 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 16 |
Hb_001053_190 |
0.1359184217 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 17 |
Hb_005535_080 |
0.136299841 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 18 |
Hb_002311_380 |
0.1388182718 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 19 |
Hb_004055_050 |
0.1412808712 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 20 |
Hb_005064_010 |
0.1422595277 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |