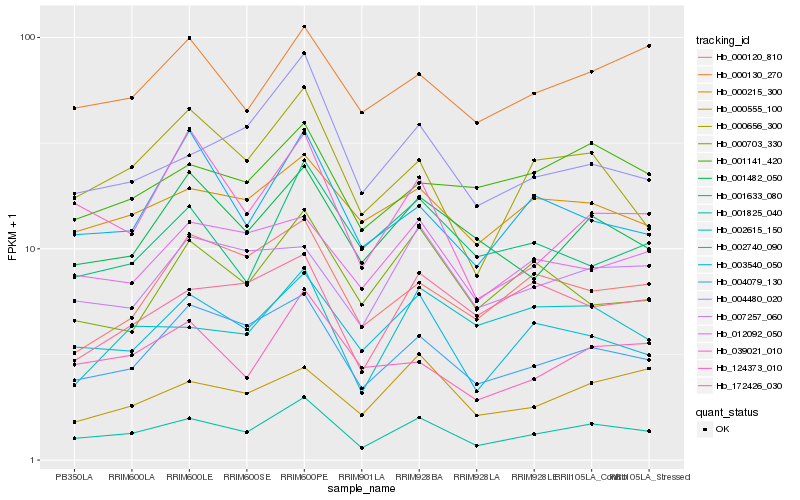
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001825_040 |
0.0 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 2 |
Hb_004079_130 |
0.0963034846 |
- |
- |
PREDICTED: uncharacterized protein LOC105637708 [Jatropha curcas] |
| 3 |
Hb_002740_090 |
0.1005383732 |
- |
- |
PREDICTED: putative membrane-bound O-acyltransferase C24H6.01c [Jatropha curcas] |
| 4 |
Hb_000656_300 |
0.1135786691 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 7 member B4-like [Populus euphratica] |
| 5 |
Hb_004480_020 |
0.1152890734 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 6 |
Hb_001482_050 |
0.1171655255 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 7 |
Hb_124373_010 |
0.1192422616 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X1 [Jatropha curcas] |
| 8 |
Hb_000215_300 |
0.1212296835 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 9 |
Hb_000130_270 |
0.1227234876 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_039021_010 |
0.1232969478 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 11 |
Hb_012092_050 |
0.124473976 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001141_420 |
0.1273358307 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003540_050 |
0.1302317977 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 14 |
Hb_172426_030 |
0.1303099939 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |
| 15 |
Hb_007257_060 |
0.1304759063 |
- |
- |
PREDICTED: omega-amidase,chloroplastic [Jatropha curcas] |
| 16 |
Hb_000120_810 |
0.131845534 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 17 |
Hb_001633_080 |
0.1323696243 |
- |
- |
PREDICTED: V-type proton ATPase subunit C [Gossypium raimondii] |
| 18 |
Hb_000703_330 |
0.1333096763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002615_150 |
0.1337501713 |
- |
- |
PREDICTED: crt homolog 1-like [Jatropha curcas] |
| 20 |
Hb_000555_100 |
0.1337786264 |
- |
- |
hypothetical protein JCGZ_14689 [Jatropha curcas] |