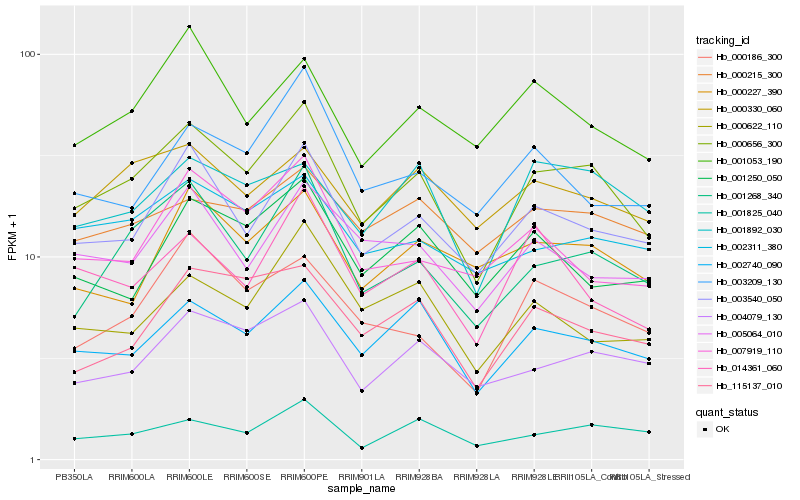
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000656_300 |
0.0 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 7 member B4-like [Populus euphratica] |
| 2 |
Hb_003540_050 |
0.0933217499 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 3 |
Hb_002740_090 |
0.0956866419 |
- |
- |
PREDICTED: putative membrane-bound O-acyltransferase C24H6.01c [Jatropha curcas] |
| 4 |
Hb_001268_340 |
0.0957587516 |
- |
- |
Actin, putative [Ricinus communis] |
| 5 |
Hb_004079_130 |
0.1081956756 |
- |
- |
PREDICTED: uncharacterized protein LOC105637708 [Jatropha curcas] |
| 6 |
Hb_001825_040 |
0.1135786691 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 7 |
Hb_002311_380 |
0.1157831204 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 8 |
Hb_000186_300 |
0.1166254941 |
- |
- |
PREDICTED: malonyl-CoA decarboxylase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000330_060 |
0.1203819959 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 10 |
Hb_115137_010 |
0.1205634624 |
- |
- |
PREDICTED: probable protein S-acyltransferase 23 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005064_010 |
0.1210052759 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 12 |
Hb_001053_190 |
0.122549401 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 13 |
Hb_014361_060 |
0.1252173334 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 14 |
Hb_000622_110 |
0.1254002872 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 15 |
Hb_007919_110 |
0.125542142 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 16 |
Hb_000227_390 |
0.1267329133 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001892_030 |
0.1267557262 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase isoform X1 [Jatropha curcas] |
| 18 |
Hb_001250_050 |
0.1270400606 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003209_130 |
0.1283246373 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 20 |
Hb_000215_300 |
0.1283280024 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |