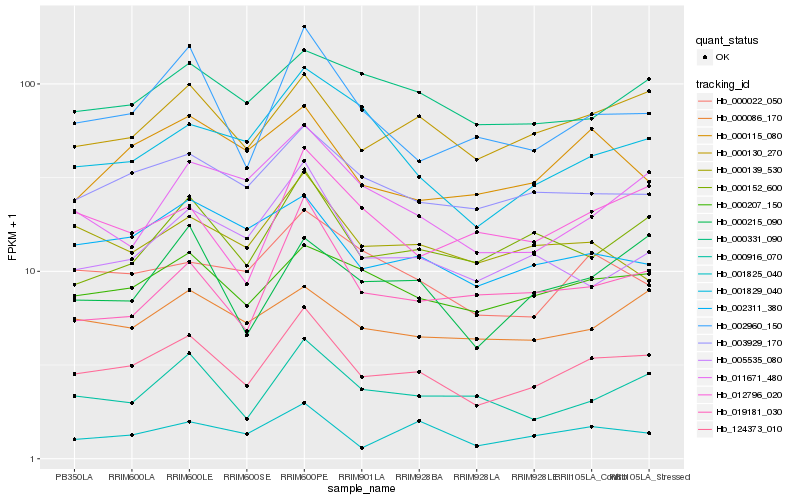
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_124373_010 |
0.0 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X1 [Jatropha curcas] |
| 2 |
Hb_000130_270 |
0.089502312 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000207_150 |
0.10200882 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 4 |
Hb_002960_150 |
0.1042251655 |
- |
- |
hypothetical protein CISIN_1g029215mg [Citrus sinensis] |
| 5 |
Hb_003929_170 |
0.1049931112 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 6 |
Hb_000152_600 |
0.1077340771 |
- |
- |
PREDICTED: protein jagunal homolog 1 [Jatropha curcas] |
| 7 |
Hb_000916_070 |
0.109108029 |
- |
- |
PREDICTED: sister chromatid cohesion protein DCC1-like [Jatropha curcas] |
| 8 |
Hb_000022_050 |
0.1140856612 |
- |
- |
PREDICTED: retinol dehydrogenase 11-like isoform X2 [Glycine max] |
| 9 |
Hb_002311_380 |
0.114929272 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 10 |
Hb_000086_170 |
0.1177767476 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 11 |
Hb_000215_090 |
0.1179266217 |
- |
- |
cytochrome b5 domain-containing family protein [Populus trichocarpa] |
| 12 |
Hb_005535_080 |
0.1184764609 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 13 |
Hb_001825_040 |
0.1192422616 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 14 |
Hb_000331_090 |
0.1196408316 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |
| 15 |
Hb_019181_030 |
0.1197346095 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 16 |
Hb_012796_020 |
0.120937317 |
- |
- |
PREDICTED: ADP-ribosylation factor 1 [Prunus mume] |
| 17 |
Hb_011671_480 |
0.1239327857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001829_040 |
0.1242183835 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 19 |
Hb_000139_530 |
0.1243726526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000115_080 |
0.1245147688 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |