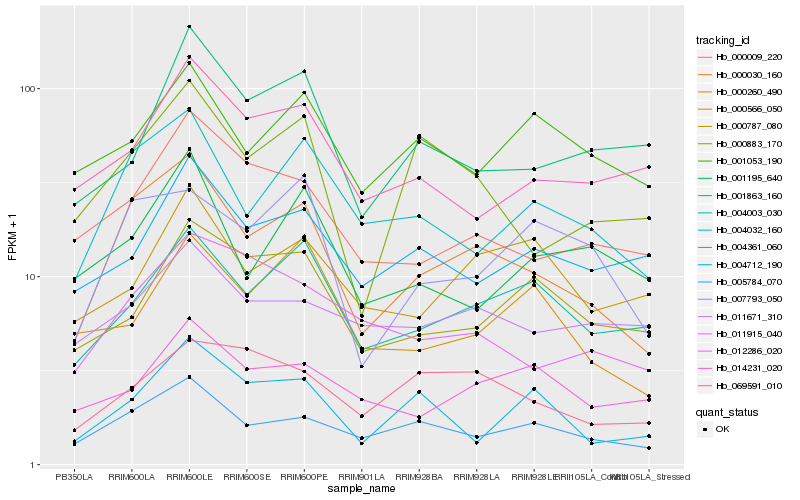
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000030_160 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 7-like [Jatropha curcas] |
| 2 |
Hb_004032_160 |
0.134415218 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 3 |
Hb_005784_070 |
0.1393685785 |
- |
- |
PREDICTED: protein TONSOKU isoform X2 [Jatropha curcas] |
| 4 |
Hb_000883_170 |
0.1650769984 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 5 |
Hb_000009_220 |
0.1657762743 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_004003_030 |
0.1683003926 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007793_050 |
0.1704166968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_011915_040 |
0.1767901208 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 9 |
Hb_069591_010 |
0.1867760649 |
- |
- |
- |
| 10 |
Hb_014231_020 |
0.1897744394 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 11 |
Hb_004361_060 |
0.1933497441 |
- |
- |
PREDICTED: uncharacterized protein LOC105636705 [Jatropha curcas] |
| 12 |
Hb_000787_080 |
0.1950178184 |
- |
- |
PREDICTED: TBC1 domain family member 16-like [Jatropha curcas] |
| 13 |
Hb_001195_640 |
0.1955898307 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 14 |
Hb_000260_490 |
0.1958658964 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 15 |
Hb_004712_190 |
0.1963313231 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 16 |
Hb_001863_160 |
0.1971176587 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 17 |
Hb_001053_190 |
0.1976551721 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 18 |
Hb_012286_020 |
0.1979455476 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 19 |
Hb_011671_310 |
0.1983790727 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 20 |
Hb_000566_050 |
0.199844086 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |