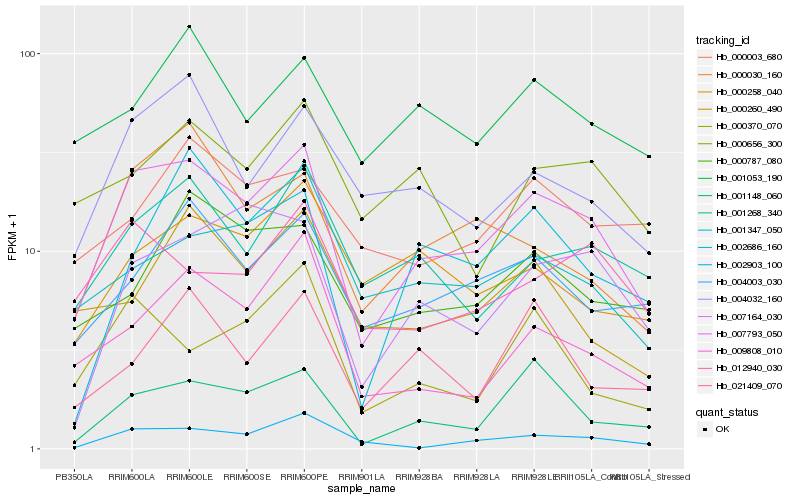
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007793_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004003_030 |
0.1604125286 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001268_340 |
0.1628914252 |
- |
- |
Actin, putative [Ricinus communis] |
| 4 |
Hb_004032_160 |
0.1637528848 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 5 |
Hb_001148_060 |
0.1649617906 |
- |
- |
hypothetical protein JCGZ_00278 [Jatropha curcas] |
| 6 |
Hb_000030_160 |
0.1704166968 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 7-like [Jatropha curcas] |
| 7 |
Hb_002903_100 |
0.1722809104 |
- |
- |
hypothetical protein POPTR_0011s04750g [Populus trichocarpa] |
| 8 |
Hb_009808_010 |
0.1744708153 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_007164_030 |
0.1759686588 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 10 |
Hb_012940_030 |
0.1779510254 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 11 |
Hb_001347_050 |
0.1782938293 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 12 |
Hb_001053_190 |
0.1793484831 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 13 |
Hb_000258_040 |
0.1805117246 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 14 |
Hb_000656_300 |
0.1827704166 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 7 member B4-like [Populus euphratica] |
| 15 |
Hb_000787_080 |
0.1829251998 |
- |
- |
PREDICTED: TBC1 domain family member 16-like [Jatropha curcas] |
| 16 |
Hb_000370_070 |
0.1876237526 |
- |
- |
PREDICTED: subtilisin-like protease-like [Citrus sinensis] |
| 17 |
Hb_000260_490 |
0.1879855964 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 18 |
Hb_002686_160 |
0.1883181789 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Populus euphratica] |
| 19 |
Hb_021409_070 |
0.1893414072 |
- |
- |
PREDICTED: transcription factor bHLH157 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000003_680 |
0.1901836905 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |