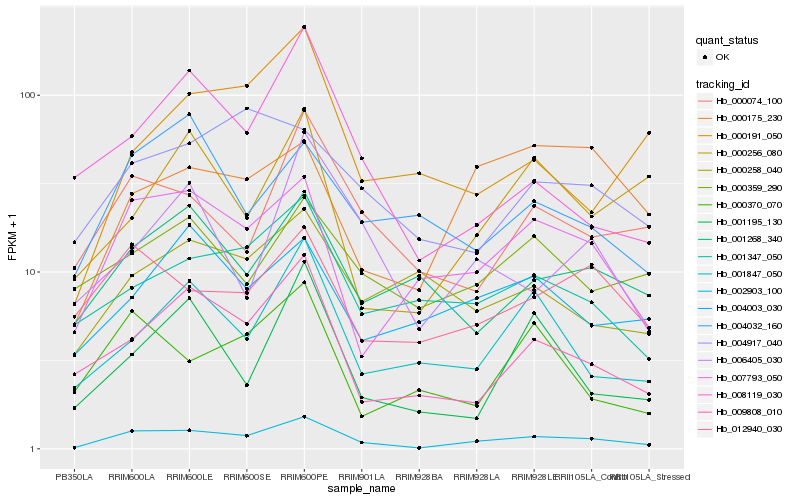
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002903_100 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s04750g [Populus trichocarpa] |
| 2 |
Hb_007793_050 |
0.1722809104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_009808_010 |
0.1846250631 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_000074_100 |
0.2023860198 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001347_050 |
0.2030791647 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 6 |
Hb_001847_050 |
0.2051074731 |
- |
- |
hypothetical protein CISIN_1g019843mg [Citrus sinensis] |
| 7 |
Hb_012940_030 |
0.2065643384 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 8 |
Hb_001268_340 |
0.2121961282 |
- |
- |
Actin, putative [Ricinus communis] |
| 9 |
Hb_008119_030 |
0.2181122566 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 10 |
Hb_004003_030 |
0.2183946179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000359_290 |
0.220806066 |
- |
- |
Triose phosphate/phosphate translocator, non-green plastid, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_001195_130 |
0.2212565243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000256_080 |
0.2230390997 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |
| 14 |
Hb_000370_070 |
0.2235017403 |
- |
- |
PREDICTED: subtilisin-like protease-like [Citrus sinensis] |
| 15 |
Hb_000175_230 |
0.2240843507 |
- |
- |
PREDICTED: protein LURP-one-related 5 [Jatropha curcas] |
| 16 |
Hb_000258_040 |
0.2245961361 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 17 |
Hb_004032_160 |
0.2255662249 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 18 |
Hb_004917_040 |
0.2273807455 |
- |
- |
hypothetical protein POPTR_0009s03470g [Populus trichocarpa] |
| 19 |
Hb_006405_030 |
0.228065751 |
- |
- |
hypothetical protein VITISV_043238 [Vitis vinifera] |
| 20 |
Hb_000191_050 |
0.2293673119 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |