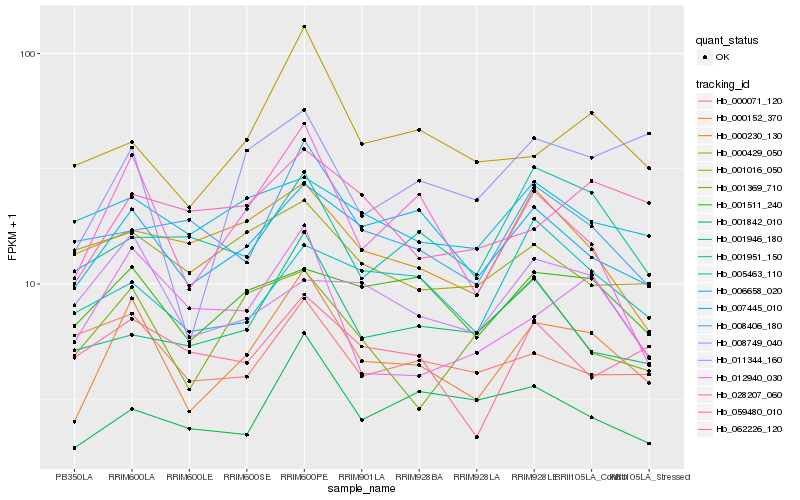
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_370 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648579 isoform X1 [Jatropha curcas] |
| 2 |
Hb_028207_060 |
0.131586206 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 3 |
Hb_007445_010 |
0.131606826 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 4 |
Hb_000429_050 |
0.1478638271 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001511_240 |
0.1487138939 |
- |
- |
PREDICTED: uncharacterized protein LOC105133574 isoform X3 [Populus euphratica] |
| 6 |
Hb_012940_030 |
0.1498490651 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 7 |
Hb_008749_040 |
0.1585005797 |
- |
- |
PREDICTED: formin-like protein 20 [Populus euphratica] |
| 8 |
Hb_001842_010 |
0.1596973987 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 9 |
Hb_001946_180 |
0.1624463235 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001369_710 |
0.1631243847 |
- |
- |
PREDICTED: uncharacterized protein LOC105648579 isoform X1 [Jatropha curcas] |
| 11 |
Hb_059480_010 |
0.1632633743 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 12 |
Hb_008406_180 |
0.1643359856 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 13 |
Hb_011344_160 |
0.1665127375 |
- |
- |
PREDICTED: primary amine oxidase [Jatropha curcas] |
| 14 |
Hb_005463_110 |
0.16751258 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase HPR3-like [Jatropha curcas] |
| 15 |
Hb_001951_150 |
0.1677765097 |
- |
- |
PREDICTED: uncharacterized protein LOC105649758 [Jatropha curcas] |
| 16 |
Hb_062226_120 |
0.1699743641 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 17 |
Hb_006658_020 |
0.1704998468 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 18 |
Hb_000230_130 |
0.1707551316 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000071_120 |
0.173061373 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 4 [Jatropha curcas] |
| 20 |
Hb_001016_050 |
0.1748477867 |
- |
- |
drought-inducible protein [Manihot esculenta] |