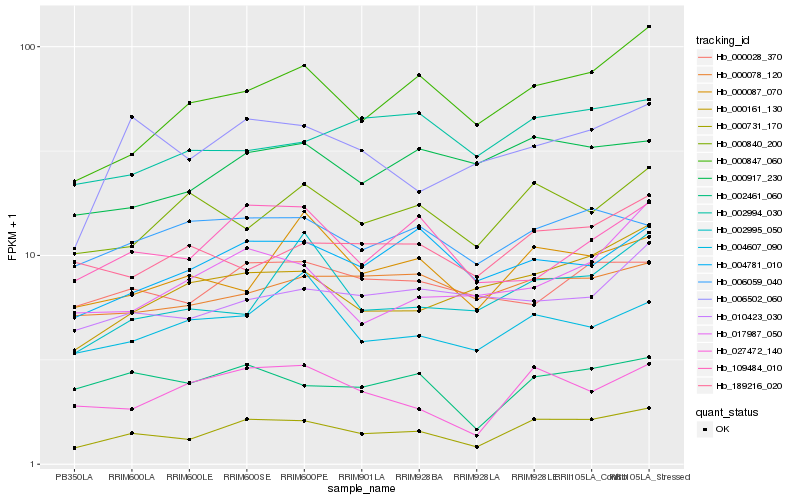
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000731_170 |
0.0 |
- |
- |
PREDICTED: phospholipase A I [Jatropha curcas] |
| 2 |
Hb_000847_060 |
0.1044426738 |
- |
- |
hypothetical protein [Jatropha curcas] |
| 3 |
Hb_000161_130 |
0.1107438614 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_027472_140 |
0.1169848639 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646176 [Jatropha curcas] |
| 5 |
Hb_000917_230 |
0.1174378625 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Jatropha curcas] |
| 6 |
Hb_002461_060 |
0.1178275194 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 7 |
Hb_004781_010 |
0.118497005 |
- |
- |
PREDICTED: potassium transporter 11-like [Populus euphratica] |
| 8 |
Hb_006502_060 |
0.1186756165 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006059_040 |
0.1255175565 |
transcription factor |
TF Family: SOH1 |
hypothetical protein PHAVU_007G063000g [Phaseolus vulgaris] |
| 10 |
Hb_000840_200 |
0.1260191827 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase small chain, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_000087_070 |
0.1264379635 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 12 |
Hb_000078_120 |
0.126850189 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 13 |
Hb_000028_370 |
0.1281810188 |
- |
- |
Guanine nucleotide-binding protein alpha-2 subunit [Glycine soja] |
| 14 |
Hb_002994_030 |
0.128974944 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-like [Pyrus x bretschneideri] |
| 15 |
Hb_010423_030 |
0.129248042 |
- |
- |
PREDICTED: uncharacterized protein LOC105645584 [Jatropha curcas] |
| 16 |
Hb_189216_020 |
0.1295845631 |
- |
- |
PREDICTED: translin [Jatropha curcas] |
| 17 |
Hb_004607_090 |
0.1295939929 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 18 |
Hb_002995_050 |
0.1298423243 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_109484_010 |
0.1300247674 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 20 |
Hb_017987_050 |
0.1312574241 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 5, chloroplastic-like [Jatropha curcas] |