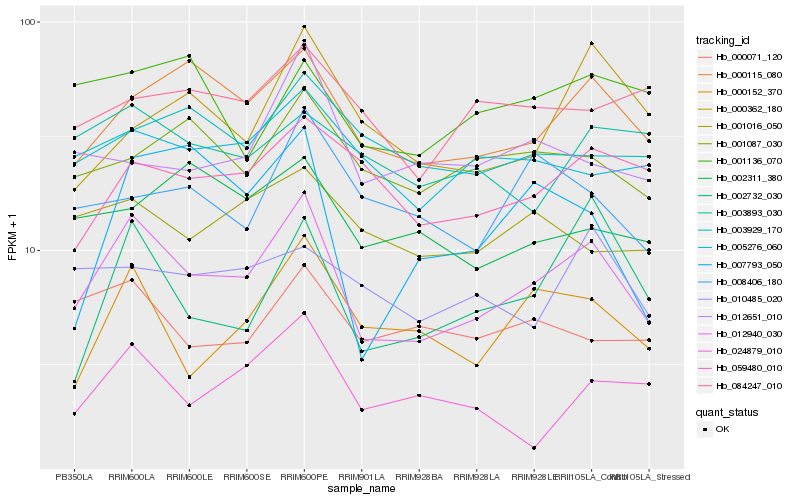
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012940_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 2 |
Hb_000115_080 |
0.1366971035 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |
| 3 |
Hb_000152_370 |
0.1498490651 |
- |
- |
PREDICTED: uncharacterized protein LOC105648579 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002732_030 |
0.1581934033 |
transcription factor |
TF Family: AP2 |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005276_060 |
0.1619400094 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_059480_010 |
0.1647428459 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 7 |
Hb_001087_030 |
0.1674070806 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 8 |
Hb_000071_120 |
0.168983223 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 4 [Jatropha curcas] |
| 9 |
Hb_024879_010 |
0.1694937683 |
- |
- |
unknown [Glycine max] |
| 10 |
Hb_001016_050 |
0.171419692 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 11 |
Hb_003929_170 |
0.1719695297 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 12 |
Hb_008406_180 |
0.1747276234 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 13 |
Hb_010485_020 |
0.1761723591 |
- |
- |
PREDICTED: thaumatin-like protein 1b isoform X1 [Jatropha curcas] |
| 14 |
Hb_012651_010 |
0.1768507351 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 15 |
Hb_000362_180 |
0.1772493438 |
- |
- |
Pdap1 [Gossypium arboreum] |
| 16 |
Hb_007793_050 |
0.1779510254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002311_380 |
0.17916575 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 18 |
Hb_001136_070 |
0.1797960098 |
- |
- |
Transmembrane protein TPARL, putative [Ricinus communis] |
| 19 |
Hb_003893_030 |
0.1803192233 |
- |
- |
Protein yrdA, putative [Ricinus communis] |
| 20 |
Hb_084247_010 |
0.1804526901 |
- |
- |
conserved hypothetical protein [Ricinus communis] |