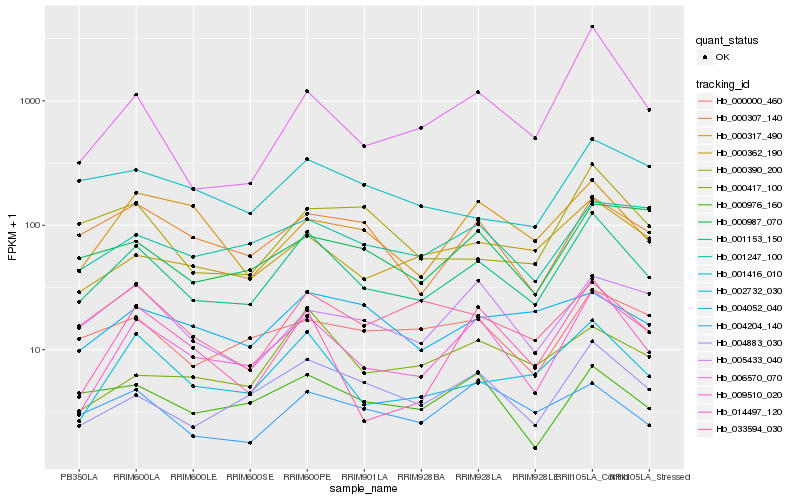
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001153_150 |
0.0 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
HMG-CoA synthase 2 [Hevea brasiliensis] |
| 2 |
Hb_009510_020 |
0.1143763479 |
- |
- |
PREDICTED: phosphatidylinositol transfer protein 2-like [Jatropha curcas] |
| 3 |
Hb_002732_030 |
0.1169188135 |
transcription factor |
TF Family: AP2 |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004883_030 |
0.1307420253 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80A2-like [Jatropha curcas] |
| 5 |
Hb_000362_190 |
0.1470346471 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 6 |
Hb_033594_030 |
0.1535236273 |
- |
- |
PREDICTED: putative glycosyltransferase 7 [Jatropha curcas] |
| 7 |
Hb_000307_140 |
0.1555302534 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2 [Jatropha curcas] |
| 8 |
Hb_000000_460 |
0.156092743 |
- |
- |
PREDICTED: peroxisomal membrane protein 11A [Jatropha curcas] |
| 9 |
Hb_000976_160 |
0.1565706931 |
- |
- |
PREDICTED: uncharacterized protein LOC105646211 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000390_200 |
0.1598052919 |
- |
- |
PREDICTED: uncharacterized protein LOC105632462 [Jatropha curcas] |
| 11 |
Hb_004204_140 |
0.1603978793 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1470470 [Ricinus communis] |
| 12 |
Hb_001416_010 |
0.1615037209 |
- |
- |
Thioredoxin H-type, putative [Ricinus communis] |
| 13 |
Hb_006570_070 |
0.1615576634 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 14 |
Hb_001247_100 |
0.1633174059 |
- |
- |
PREDICTED: pyroglutamyl-peptidase 1-like protein [Jatropha curcas] |
| 15 |
Hb_005433_040 |
0.1653395699 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 16 |
Hb_014497_120 |
0.1687436936 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 1; Short=HMG-CoA reductase 1 [Hevea brasiliensis] |
| 17 |
Hb_004052_040 |
0.1689355569 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 18 |
Hb_000417_100 |
0.1690788698 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 12-like [Jatropha curcas] |
| 19 |
Hb_000317_490 |
0.1714977767 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 20 |
Hb_000987_070 |
0.1721541445 |
- |
- |
Inflorescence meristem receptor-like kinase 2 isoform 1 [Theobroma cacao] |