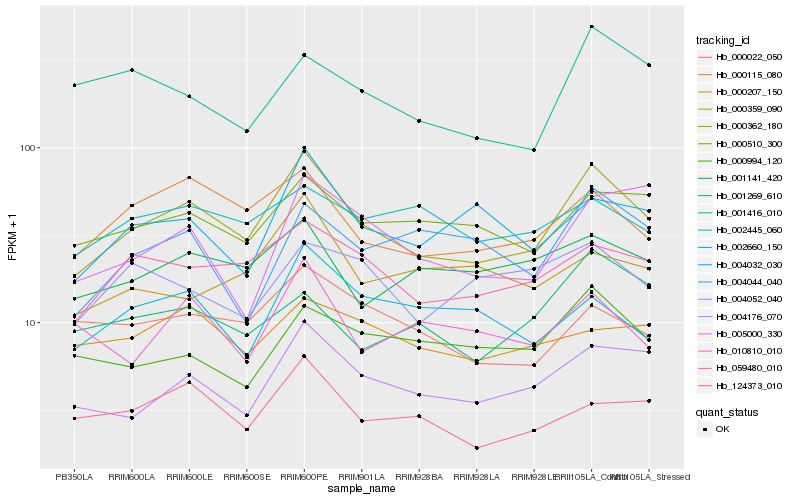
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000362_180 |
0.0 |
- |
- |
Pdap1 [Gossypium arboreum] |
| 2 |
Hb_004176_070 |
0.1125245432 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_059480_010 |
0.1152604881 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 4 |
Hb_000115_080 |
0.1190034739 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |
| 5 |
Hb_000510_300 |
0.1283442935 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 6 |
Hb_004032_030 |
0.1312562566 |
- |
- |
PREDICTED: RNA-binding protein 24-like isoform X1 [Populus euphratica] |
| 7 |
Hb_124373_010 |
0.132142228 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X1 [Jatropha curcas] |
| 8 |
Hb_001141_420 |
0.1371419926 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000022_050 |
0.138231097 |
- |
- |
PREDICTED: retinol dehydrogenase 11-like isoform X2 [Glycine max] |
| 10 |
Hb_005000_330 |
0.1433519691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000994_120 |
0.1454499545 |
- |
- |
unknown [Populus trichocarpa] |
| 12 |
Hb_002660_150 |
0.1457701341 |
- |
- |
PREDICTED: uncharacterized protein LOC105642207 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002445_060 |
0.1476373697 |
- |
- |
arf gtpase-activating protein, putative [Ricinus communis] |
| 14 |
Hb_010810_010 |
0.1483333449 |
- |
- |
PREDICTED: plastidic glucose transporter 4 [Jatropha curcas] |
| 15 |
Hb_001416_010 |
0.1494378426 |
- |
- |
Thioredoxin H-type, putative [Ricinus communis] |
| 16 |
Hb_004044_040 |
0.1494921027 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 17 |
Hb_000359_090 |
0.1509583842 |
- |
- |
PREDICTED: persulfide dioxygenase ETHE1 homolog, mitochondrial-like [Malus domestica] |
| 18 |
Hb_004052_040 |
0.1519768309 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 19 |
Hb_001269_610 |
0.1523242346 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_000207_150 |
0.1525543289 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |