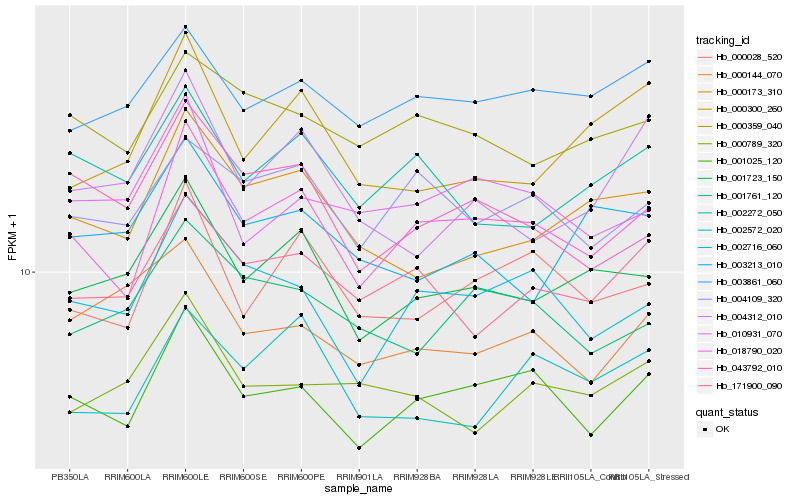
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001723_150 |
0.0 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_003213_010 |
0.0616914314 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 3 |
Hb_000173_310 |
0.0639969416 |
- |
- |
PREDICTED: uncharacterized protein LOC105631893 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000300_260 |
0.0719495031 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 5 |
Hb_002272_050 |
0.0738477523 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 6 |
Hb_003861_060 |
0.0747223515 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 7 |
Hb_018790_020 |
0.0789973265 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000028_520 |
0.0792564796 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 9 |
Hb_043792_010 |
0.082414663 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X2 [Jatropha curcas] |
| 10 |
Hb_171900_090 |
0.0830768656 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000359_040 |
0.0834495958 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004312_010 |
0.0848110568 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001761_120 |
0.0863112002 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 14 |
Hb_000789_320 |
0.0881636587 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_010931_070 |
0.0883712432 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000144_070 |
0.0883997147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004109_320 |
0.0884837872 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 18 |
Hb_001025_120 |
0.0886320231 |
transcription factor |
TF Family: Jumonji |
PREDICTED: jmjC domain-containing protein 4 [Jatropha curcas] |
| 19 |
Hb_002716_060 |
0.088775115 |
- |
- |
carboxylesterase np, putative [Ricinus communis] |
| 20 |
Hb_002572_020 |
0.0898828688 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |