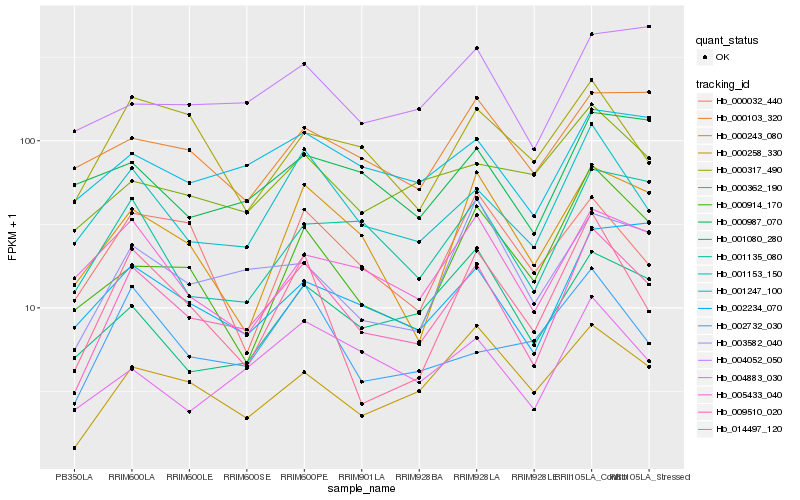
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009510_020 |
0.0 |
- |
- |
PREDICTED: phosphatidylinositol transfer protein 2-like [Jatropha curcas] |
| 2 |
Hb_001153_150 |
0.1143763479 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
HMG-CoA synthase 2 [Hevea brasiliensis] |
| 3 |
Hb_000258_330 |
0.139872118 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 4 |
Hb_003582_040 |
0.1480670532 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0010s11160g [Populus trichocarpa] |
| 5 |
Hb_014497_120 |
0.1481480377 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 1; Short=HMG-CoA reductase 1 [Hevea brasiliensis] |
| 6 |
Hb_004883_030 |
0.1486016531 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80A2-like [Jatropha curcas] |
| 7 |
Hb_001247_100 |
0.1507679315 |
- |
- |
PREDICTED: pyroglutamyl-peptidase 1-like protein [Jatropha curcas] |
| 8 |
Hb_000243_080 |
0.1511476118 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |
| 9 |
Hb_001135_080 |
0.1561474101 |
- |
- |
PREDICTED: CDP-diacylglycerol--glycerol-3-phosphate 3-phosphatidyltransferase 2-like [Jatropha curcas] |
| 10 |
Hb_005433_040 |
0.1599761141 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 11 |
Hb_001080_280 |
0.160651484 |
- |
- |
PREDICTED: uncharacterized protein LOC105649981 [Jatropha curcas] |
| 12 |
Hb_002732_030 |
0.1609945345 |
transcription factor |
TF Family: AP2 |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002234_070 |
0.1638905288 |
- |
- |
PREDICTED: polynucleotide 5'-hydroxyl-kinase nol9 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000914_170 |
0.1642404293 |
- |
- |
hypothetical protein JCGZ_25909 [Jatropha curcas] |
| 15 |
Hb_000362_190 |
0.1654970602 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 16 |
Hb_000317_490 |
0.1672653867 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 17 |
Hb_000103_320 |
0.1721459534 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 18 |
Hb_000032_440 |
0.1723483742 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_000987_070 |
0.1725003409 |
- |
- |
Inflorescence meristem receptor-like kinase 2 isoform 1 [Theobroma cacao] |
| 20 |
Hb_004052_050 |
0.1731658279 |
- |
- |
PREDICTED: uncharacterized protein LOC105640640 [Jatropha curcas] |