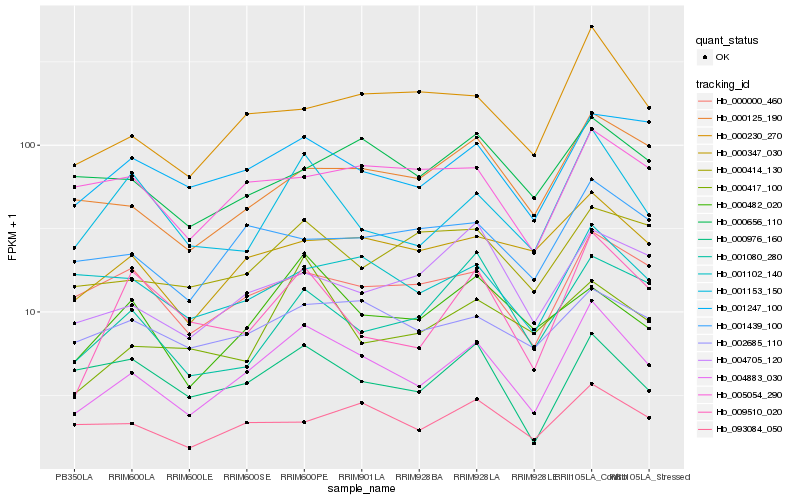
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004883_030 |
0.0 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80A2-like [Jatropha curcas] |
| 2 |
Hb_001153_150 |
0.1307420253 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
HMG-CoA synthase 2 [Hevea brasiliensis] |
| 3 |
Hb_000230_270 |
0.1334900175 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 4 |
Hb_000347_030 |
0.1387710803 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 5 |
Hb_000125_190 |
0.1402039772 |
- |
- |
PREDICTED: F-box protein SKIP16 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001247_100 |
0.1404734898 |
- |
- |
PREDICTED: pyroglutamyl-peptidase 1-like protein [Jatropha curcas] |
| 7 |
Hb_000000_460 |
0.1426957209 |
- |
- |
PREDICTED: peroxisomal membrane protein 11A [Jatropha curcas] |
| 8 |
Hb_009510_020 |
0.1486016531 |
- |
- |
PREDICTED: phosphatidylinositol transfer protein 2-like [Jatropha curcas] |
| 9 |
Hb_093084_050 |
0.1487551048 |
- |
- |
PREDICTED: uncharacterized protein LOC105640739 [Jatropha curcas] |
| 10 |
Hb_001102_140 |
0.1488992113 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 2-like [Jatropha curcas] |
| 11 |
Hb_001439_100 |
0.1501708088 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 12 |
Hb_000414_130 |
0.1540405447 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 13 |
Hb_000482_020 |
0.1544171672 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 14 |
Hb_002685_110 |
0.1553973379 |
- |
- |
BTB/POZ domain-containing protein KCTD9, putative [Ricinus communis] |
| 15 |
Hb_004705_120 |
0.1576212004 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_005054_290 |
0.1580820391 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit F [Jatropha curcas] |
| 17 |
Hb_000417_100 |
0.1587987143 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 12-like [Jatropha curcas] |
| 18 |
Hb_001080_280 |
0.1595132168 |
- |
- |
PREDICTED: uncharacterized protein LOC105649981 [Jatropha curcas] |
| 19 |
Hb_000976_160 |
0.1595652178 |
- |
- |
PREDICTED: uncharacterized protein LOC105646211 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000656_110 |
0.1598088608 |
- |
- |
nucleolysin tia-1, putative [Ricinus communis] |