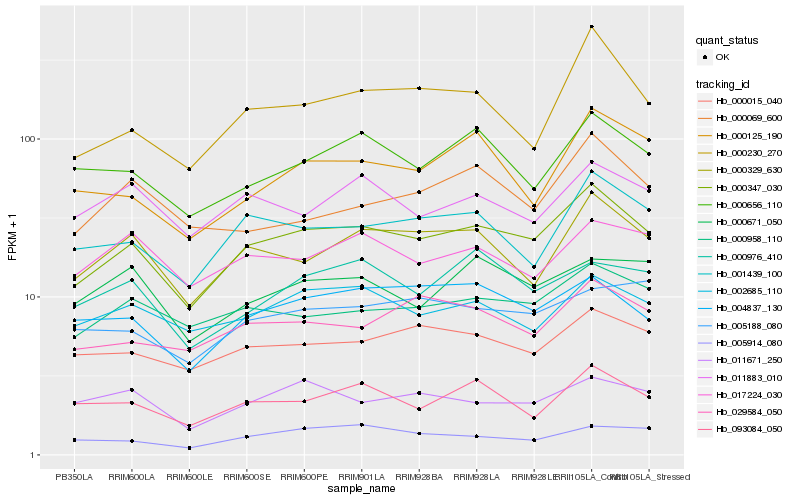
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_030 |
0.0 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 2 |
Hb_000329_630 |
0.0950723606 |
- |
- |
PREDICTED: nuclear pore complex protein NUP62 [Jatropha curcas] |
| 3 |
Hb_001439_100 |
0.0955747414 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 4 |
Hb_000958_110 |
0.0970709689 |
- |
- |
PREDICTED: uncharacterized protein LOC105628719 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000015_040 |
0.101485952 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004837_130 |
0.1033737544 |
- |
- |
PREDICTED: armadillo repeat-containing protein 8 [Jatropha curcas] |
| 7 |
Hb_029584_050 |
0.1045659252 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3 [Jatropha curcas] |
| 8 |
Hb_000230_270 |
0.1060447818 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 9 |
Hb_000656_110 |
0.1067344028 |
- |
- |
nucleolysin tia-1, putative [Ricinus communis] |
| 10 |
Hb_002685_110 |
0.1077021451 |
- |
- |
BTB/POZ domain-containing protein KCTD9, putative [Ricinus communis] |
| 11 |
Hb_093084_050 |
0.1088060979 |
- |
- |
PREDICTED: uncharacterized protein LOC105640739 [Jatropha curcas] |
| 12 |
Hb_000125_190 |
0.1131625061 |
- |
- |
PREDICTED: F-box protein SKIP16 isoform X2 [Jatropha curcas] |
| 13 |
Hb_017224_030 |
0.1176304175 |
- |
- |
PREDICTED: A-agglutinin anchorage subunit [Jatropha curcas] |
| 14 |
Hb_011883_010 |
0.1178511852 |
- |
- |
PREDICTED: RNA-binding protein 7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005188_080 |
0.117964469 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 16 |
Hb_011671_250 |
0.1183931515 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_000976_410 |
0.1186172173 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 18 |
Hb_005914_080 |
0.1200464971 |
- |
- |
PREDICTED: uncharacterized protein LOC105632176 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000671_050 |
0.1202120307 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 20 |
Hb_000069_600 |
0.120918536 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |