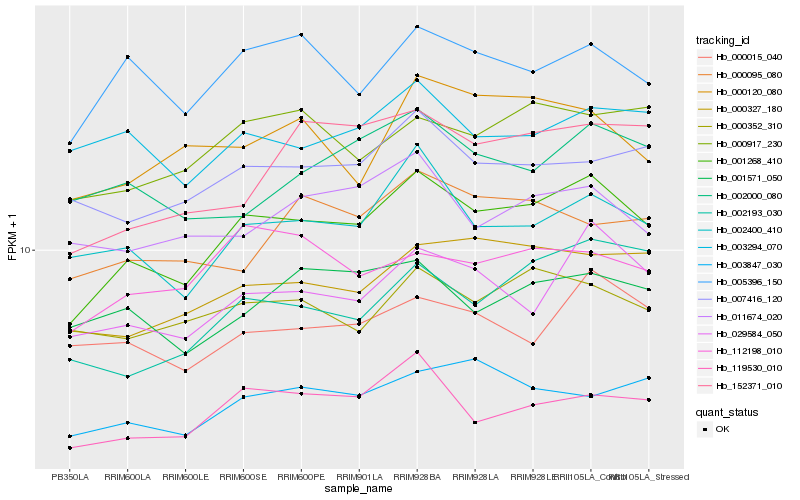
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001268_410 |
0.0 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP20 [Jatropha curcas] |
| 2 |
Hb_029584_050 |
0.0882185833 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3 [Jatropha curcas] |
| 3 |
Hb_002400_410 |
0.0889990422 |
- |
- |
PREDICTED: topless-related protein 1-like [Jatropha curcas] |
| 4 |
Hb_119530_010 |
0.0897026849 |
- |
- |
- |
| 5 |
Hb_000327_180 |
0.0931349717 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_005396_150 |
0.0954644568 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_000917_230 |
0.0997099039 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Jatropha curcas] |
| 8 |
Hb_007416_120 |
0.1020858939 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 9 |
Hb_000352_310 |
0.1025205009 |
- |
- |
hypothetical protein CISIN_1g0017891mg, partial [Citrus sinensis] |
| 10 |
Hb_152371_010 |
0.1025715778 |
- |
- |
RNA-binding KH domain-containing protein [Theobroma cacao] |
| 11 |
Hb_003847_030 |
0.1033040248 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15010, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000015_040 |
0.1034714988 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001571_050 |
0.1053962992 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3-like [Populus euphratica] |
| 14 |
Hb_112198_010 |
0.1059423305 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |
| 15 |
Hb_002000_080 |
0.1068397816 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002193_030 |
0.1069457811 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like [Jatropha curcas] |
| 17 |
Hb_011674_020 |
0.1070507326 |
- |
- |
PREDICTED: NEDD8-activating enzyme E1 regulatory subunit-like [Jatropha curcas] |
| 18 |
Hb_000120_080 |
0.1070902046 |
- |
- |
PREDICTED: probable pyridoxal 5'-phosphate synthase subunit PDX1 [Jatropha curcas] |
| 19 |
Hb_003294_070 |
0.1097877817 |
- |
- |
hypothetical protein RCOM_1714550 [Ricinus communis] |
| 20 |
Hb_000095_080 |
0.11118291 |
- |
- |
PREDICTED: probable U3 small nucleolar RNA-associated protein 7 [Jatropha curcas] |