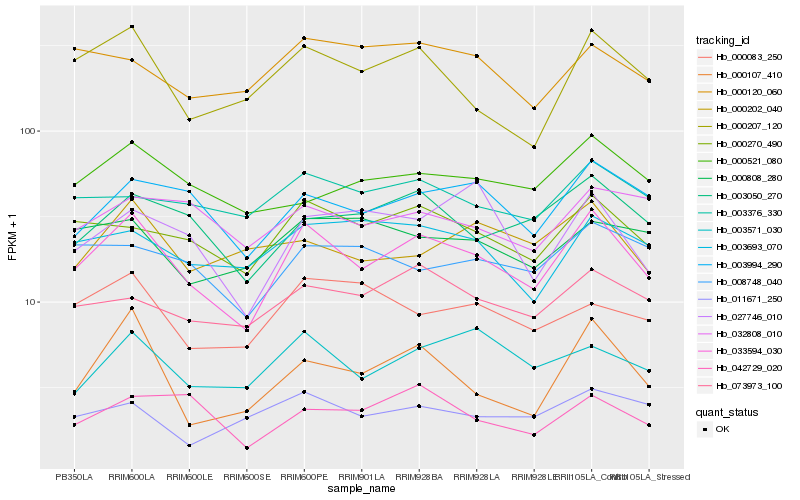
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033594_030 |
0.0 |
- |
- |
PREDICTED: putative glycosyltransferase 7 [Jatropha curcas] |
| 2 |
Hb_000270_490 |
0.111959015 |
- |
- |
betaine aldehyde dehydrogenase 1 [Jatropha curcas] |
| 3 |
Hb_000207_120 |
0.1158668109 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.3 [Jatropha curcas] |
| 4 |
Hb_003994_290 |
0.120664014 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase GAPCP2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003050_270 |
0.1241433302 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase I, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003571_030 |
0.1284320676 |
- |
- |
hypothetical protein JCGZ_25894 [Jatropha curcas] |
| 7 |
Hb_073973_100 |
0.1306291771 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |
| 8 |
Hb_003376_330 |
0.1354149552 |
- |
- |
PREDICTED: gamma carbonic anhydrase 1, mitochondrial [Eucalyptus grandis] |
| 9 |
Hb_003693_070 |
0.1359031483 |
- |
- |
PREDICTED: S-(hydroxymethyl)glutathione dehydrogenase [Jatropha curcas] |
| 10 |
Hb_000107_410 |
0.136900944 |
- |
- |
PREDICTED: expansin-A20 [Jatropha curcas] |
| 11 |
Hb_000521_080 |
0.138088898 |
- |
- |
Zinc finger BED domain-containing 4 [Gossypium arboreum] |
| 12 |
Hb_000083_250 |
0.1393987137 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 13 |
Hb_008748_040 |
0.1420825773 |
- |
- |
beta-hydroxyacyl-acyl carrier protein dehydratase [Hevea brasiliensis] |
| 14 |
Hb_000120_060 |
0.1428102163 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP20-1 [Populus euphratica] |
| 15 |
Hb_042729_020 |
0.1430063287 |
- |
- |
PREDICTED: protein argonaute 16 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000808_280 |
0.1434399559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_011671_250 |
0.1438450423 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_032808_010 |
0.1448529195 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 19 |
Hb_000202_040 |
0.145034523 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BOI-like [Jatropha curcas] |
| 20 |
Hb_027746_010 |
0.1455216123 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase GDPD6 isoform X1 [Jatropha curcas] |