| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
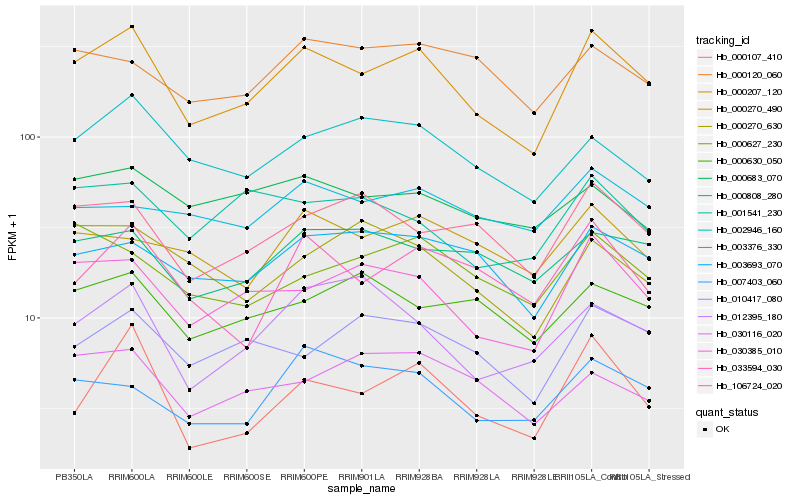
Hb_000207_120 |
0.0 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.3 [Jatropha curcas] |
| 2 |
Hb_030385_010 |
0.0979126828 |
- |
- |
PREDICTED: zinc-binding alcohol dehydrogenase domain-containing protein 2-like [Jatropha curcas] |
| 3 |
Hb_003693_070 |
0.1131284127 |
- |
- |
PREDICTED: S-(hydroxymethyl)glutathione dehydrogenase [Jatropha curcas] |
| 4 |
Hb_002946_160 |
0.1134255857 |
- |
- |
PREDICTED: uricase-2 isozyme 2 [Jatropha curcas] |
| 5 |
Hb_033594_030 |
0.1158668109 |
- |
- |
PREDICTED: putative glycosyltransferase 7 [Jatropha curcas] |
| 6 |
Hb_106724_020 |
0.1191505667 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 1A-like isoform X1 [Populus euphratica] |
| 7 |
Hb_000808_280 |
0.1209188916 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_010417_080 |
0.1213025481 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007403_060 |
0.1221022019 |
- |
- |
PREDICTED: CAAX prenyl protease 2 [Jatropha curcas] |
| 10 |
Hb_030116_020 |
0.1272957842 |
- |
- |
hypothetical protein POPTR_0016s13630g [Populus trichocarpa] |
| 11 |
Hb_000270_490 |
0.1274603392 |
- |
- |
betaine aldehyde dehydrogenase 1 [Jatropha curcas] |
| 12 |
Hb_000120_060 |
0.1284006912 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP20-1 [Populus euphratica] |
| 13 |
Hb_000683_070 |
0.1288489102 |
- |
- |
d-3-phosphoglycerate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_001541_230 |
0.1289400385 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000630_050 |
0.1297744206 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000627_230 |
0.1297827753 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000270_630 |
0.1304039042 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like [Jatropha curcas] |
| 18 |
Hb_000107_410 |
0.1315109912 |
- |
- |
PREDICTED: expansin-A20 [Jatropha curcas] |
| 19 |
Hb_012395_180 |
0.132266544 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 20 |
Hb_003376_330 |
0.1323988816 |
- |
- |
PREDICTED: gamma carbonic anhydrase 1, mitochondrial [Eucalyptus grandis] |