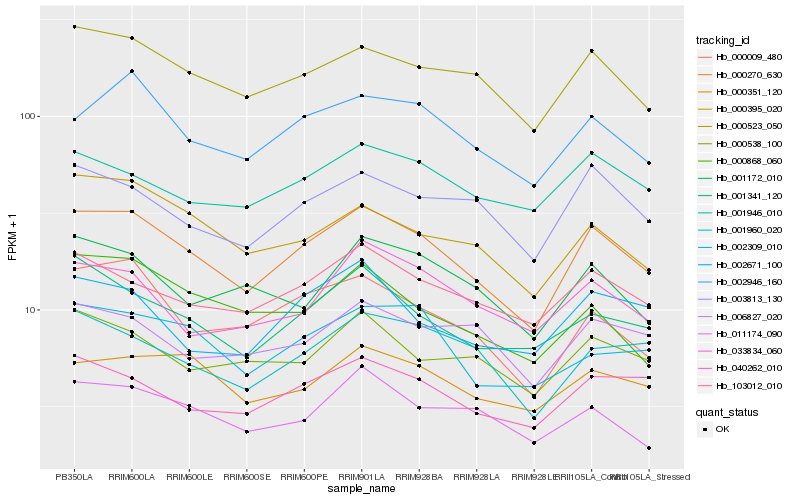
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_630 |
0.0 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like [Jatropha curcas] |
| 2 |
Hb_000523_050 |
0.0733710419 |
- |
- |
microsomal glutathione s-transferase, putative [Ricinus communis] |
| 3 |
Hb_002946_160 |
0.0830505375 |
- |
- |
PREDICTED: uricase-2 isozyme 2 [Jatropha curcas] |
| 4 |
Hb_000009_480 |
0.0840119689 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 5 |
Hb_103012_010 |
0.0851230261 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 6 |
Hb_003813_130 |
0.0856315387 |
- |
- |
PREDICTED: plastidic glucose transporter 4 [Jatropha curcas] |
| 7 |
Hb_040262_010 |
0.088367695 |
- |
- |
PREDICTED: aspartate carbamoyltransferase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000395_020 |
0.089203182 |
- |
- |
PREDICTED: ankyrin repeat and KH domain-containing protein R11A8.7 [Jatropha curcas] |
| 9 |
Hb_001341_120 |
0.0900213693 |
- |
- |
hypothetical protein B456_001G130300 [Gossypium raimondii] |
| 10 |
Hb_000351_120 |
0.0901447077 |
- |
- |
hypothetical protein POPTR_0013s10290g [Populus trichocarpa] |
| 11 |
Hb_002309_010 |
0.0918085367 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 12 |
Hb_001960_020 |
0.0926435005 |
- |
- |
PREDICTED: mitochondrial arginine transporter BAC2 [Jatropha curcas] |
| 13 |
Hb_000538_100 |
0.0940591553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_011174_090 |
0.095087297 |
- |
- |
PREDICTED: fidgetin-like protein 1 [Jatropha curcas] |
| 15 |
Hb_001172_010 |
0.0970571642 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 16 |
Hb_001946_010 |
0.0972898448 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 17 |
Hb_006827_020 |
0.0976192489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_033834_060 |
0.0978128778 |
- |
- |
DNA-binding bromodomain-containing family protein [Populus trichocarpa] |
| 19 |
Hb_002671_100 |
0.0993977424 |
- |
- |
PREDICTED: tRNA (guanine-N(7)-)-methyltransferase-like [Jatropha curcas] |
| 20 |
Hb_000868_060 |
0.0996676291 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |