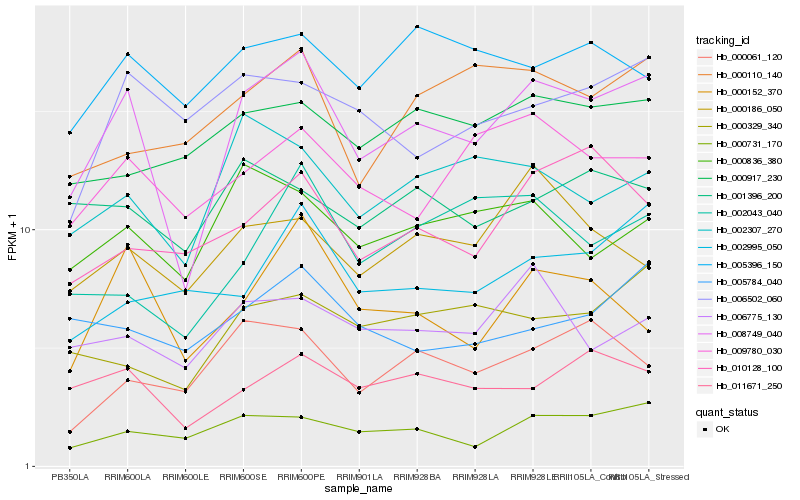
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008749_040 |
0.0 |
- |
- |
PREDICTED: formin-like protein 20 [Populus euphratica] |
| 2 |
Hb_011671_250 |
0.1455197022 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_000731_170 |
0.1492066041 |
- |
- |
PREDICTED: phospholipase A I [Jatropha curcas] |
| 4 |
Hb_002307_270 |
0.155987815 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 5 |
Hb_009780_030 |
0.1568883885 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 6 |
Hb_000152_370 |
0.1585005797 |
- |
- |
PREDICTED: uncharacterized protein LOC105648579 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000836_380 |
0.1595299593 |
- |
- |
PREDICTED: C-terminal binding protein AN-like [Populus euphratica] |
| 8 |
Hb_006775_130 |
0.1635243721 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 9 |
Hb_000329_340 |
0.1638470537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002043_040 |
0.1685565 |
- |
- |
PREDICTED: uncharacterized protein LOC105631509 [Jatropha curcas] |
| 11 |
Hb_000110_140 |
0.16999686 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 12 |
Hb_000917_230 |
0.170410419 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Jatropha curcas] |
| 13 |
Hb_006502_060 |
0.1706102556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000186_050 |
0.1711247698 |
- |
- |
PREDICTED: uncharacterized protein LOC105649281 [Jatropha curcas] |
| 15 |
Hb_010128_100 |
0.1715025245 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000061_120 |
0.1717358557 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 16 [Jatropha curcas] |
| 17 |
Hb_005784_040 |
0.1717883876 |
- |
- |
PREDICTED: uncharacterized protein LOC105645160 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005396_150 |
0.1724844647 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_001396_200 |
0.1726540495 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002995_050 |
0.1727265872 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |