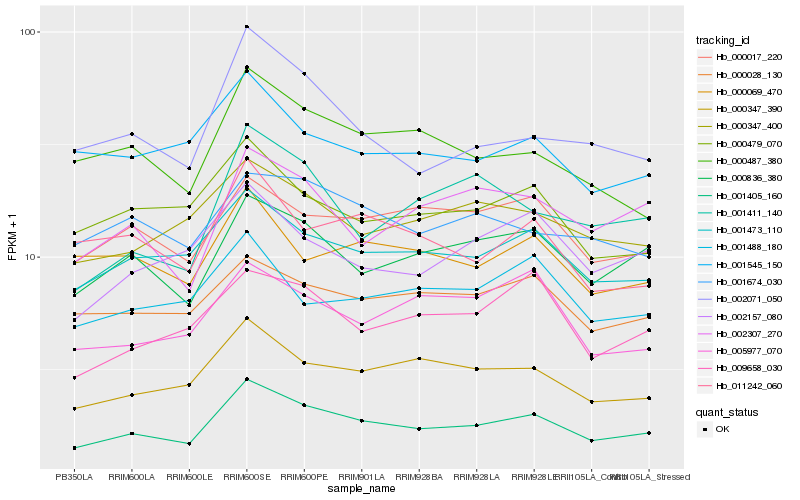
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001405_160 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g09680 [Jatropha curcas] |
| 2 |
Hb_001473_110 |
0.0747011701 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 3 |
Hb_000347_390 |
0.0748790808 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 4 |
Hb_000836_380 |
0.0749524234 |
- |
- |
PREDICTED: C-terminal binding protein AN-like [Populus euphratica] |
| 5 |
Hb_000347_400 |
0.090639105 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000017_220 |
0.0907668415 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 7 |
Hb_002307_270 |
0.0934631726 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 8 |
Hb_009658_030 |
0.0940004593 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 9 |
Hb_000028_130 |
0.0945995446 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000479_070 |
0.0955926784 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 11 |
Hb_000487_380 |
0.0957046768 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002157_080 |
0.0959988044 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002071_050 |
0.0973491657 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 5-like [Jatropha curcas] |
| 14 |
Hb_001674_030 |
0.0984422762 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g24240 [Jatropha curcas] |
| 15 |
Hb_001411_140 |
0.0991045648 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 16 |
Hb_005977_070 |
0.1003394639 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001488_180 |
0.1011679892 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 18 |
Hb_011242_060 |
0.1019700055 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 19 |
Hb_000069_470 |
0.1027781434 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001545_150 |
0.1028341627 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |