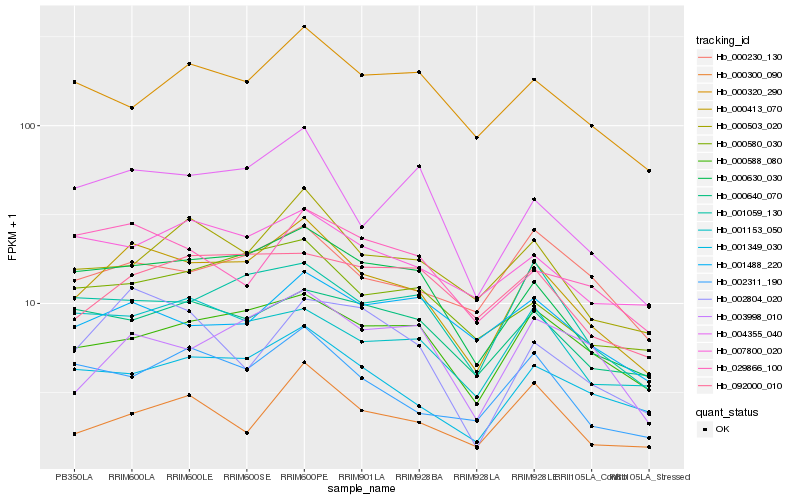
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000413_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000630_030 |
0.0959365417 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 isoform X1 [Populus euphratica] |
| 3 |
Hb_001349_030 |
0.1133714698 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004355_040 |
0.1142307087 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 5 |
Hb_000588_080 |
0.1151712451 |
- |
- |
PREDICTED: uncharacterized protein LOC105638459 [Jatropha curcas] |
| 6 |
Hb_001059_130 |
0.1205976427 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000580_030 |
0.1223306889 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 8 |
Hb_000503_020 |
0.122844221 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 9 |
Hb_002311_190 |
0.1277542352 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 10 |
Hb_003998_010 |
0.1278303275 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 11 |
Hb_000640_070 |
0.1323242519 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 12 |
Hb_000230_130 |
0.1324534942 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007800_020 |
0.1341584532 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_092000_010 |
0.1356358937 |
- |
- |
kelch repeat-containing F-box family protein [Populus trichocarpa] |
| 15 |
Hb_001153_050 |
0.137011211 |
- |
- |
PREDICTED: protein FIZZY-RELATED 2 [Jatropha curcas] |
| 16 |
Hb_029866_100 |
0.1383810955 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 17 |
Hb_000320_290 |
0.1387767173 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 18 |
Hb_002804_020 |
0.1389490905 |
- |
- |
epsin N-terminal homology domain-containing family protein [Populus trichocarpa] |
| 19 |
Hb_001488_220 |
0.1396455623 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 20 |
Hb_000300_090 |
0.1406864806 |
- |
- |
catalytic, putative [Ricinus communis] |