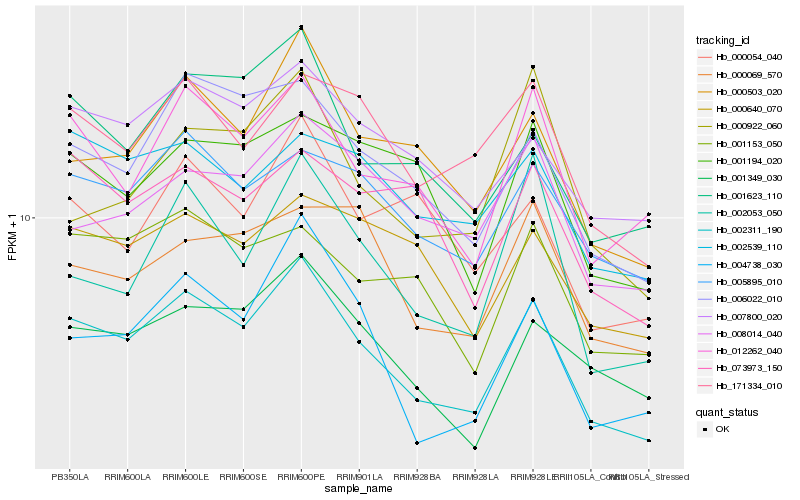
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 2 |
Hb_006022_010 |
0.090994037 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 3 |
Hb_012262_040 |
0.0984785121 |
- |
- |
PREDICTED: uncharacterized protein LOC105633697 [Jatropha curcas] |
| 4 |
Hb_000503_020 |
0.0988800719 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 5 |
Hb_001623_110 |
0.1006410581 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_004738_030 |
0.1022021125 |
- |
- |
Inactive protein kinase [Gossypium arboreum] |
| 7 |
Hb_001349_030 |
0.1050895435 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007800_020 |
0.1060027138 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005895_010 |
0.109695668 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 10 |
Hb_171334_010 |
0.1107660585 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 11 |
Hb_001153_050 |
0.1130551081 |
- |
- |
PREDICTED: protein FIZZY-RELATED 2 [Jatropha curcas] |
| 12 |
Hb_002053_050 |
0.1141302839 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 13 |
Hb_000069_570 |
0.1197095974 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002539_110 |
0.1198838677 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 15 |
Hb_000054_040 |
0.121149866 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 16 |
Hb_001194_020 |
0.1218788762 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 17 |
Hb_000640_070 |
0.1232772793 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 18 |
Hb_008014_040 |
0.1237500277 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 19 |
Hb_073973_150 |
0.1240187104 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000922_060 |
0.1268913904 |
- |
- |
kinesin light chain, putative [Ricinus communis] |