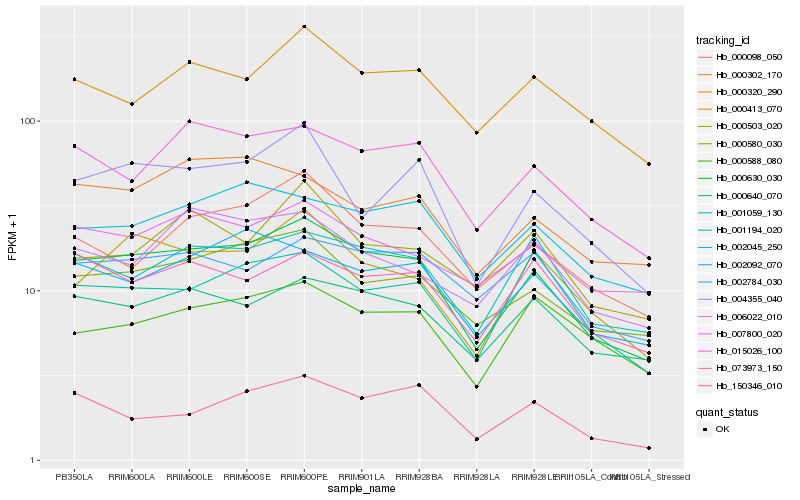
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000630_030 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 isoform X1 [Populus euphratica] |
| 2 |
Hb_000580_030 |
0.0841793667 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 3 |
Hb_015026_100 |
0.093108218 |
- |
- |
PREDICTED: transmembrane protein 184C-like [Jatropha curcas] |
| 4 |
Hb_000640_070 |
0.0938128099 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 5 |
Hb_000413_070 |
0.0959365417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004355_040 |
0.0977920717 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 7 |
Hb_002092_070 |
0.0999678738 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 8 |
Hb_001194_020 |
0.1003900155 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 9 |
Hb_002045_250 |
0.1009864443 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000098_050 |
0.1019158208 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 11 |
Hb_007800_020 |
0.1029273213 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000320_290 |
0.1041410299 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 13 |
Hb_073973_150 |
0.1072130045 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002784_030 |
0.1076495357 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 15 |
Hb_006022_010 |
0.1146584469 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 16 |
Hb_001059_130 |
0.11468605 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000302_170 |
0.1147749918 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 18 |
Hb_150346_010 |
0.1149355923 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 19 |
Hb_000588_080 |
0.1154291304 |
- |
- |
PREDICTED: uncharacterized protein LOC105638459 [Jatropha curcas] |
| 20 |
Hb_000503_020 |
0.116561827 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |