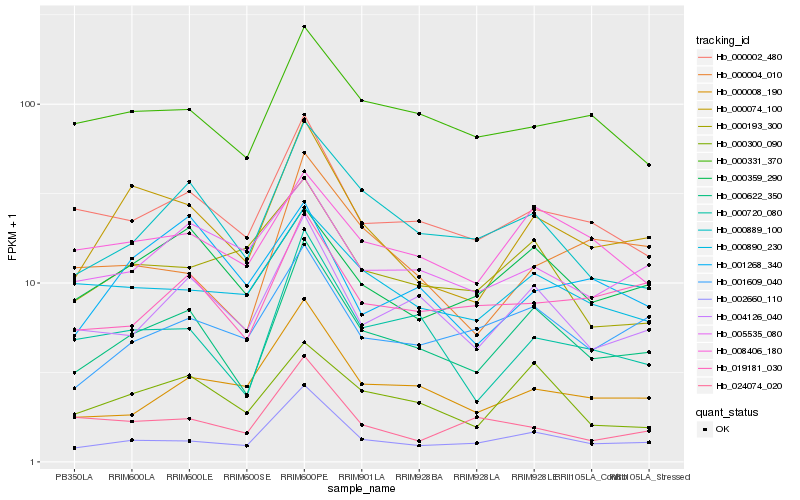
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000074_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000622_350 |
0.1154709126 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 3 |
Hb_002660_110 |
0.1325356684 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000359_290 |
0.1573154712 |
- |
- |
Triose phosphate/phosphate translocator, non-green plastid, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_000002_480 |
0.1575269702 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000890_230 |
0.1605762562 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 7 |
Hb_000720_080 |
0.1639515631 |
- |
- |
PREDICTED: bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1 [Jatropha curcas] |
| 8 |
Hb_000004_010 |
0.1648135812 |
- |
- |
hypothetical protein Csa_6G120400 [Cucumis sativus] |
| 9 |
Hb_000331_370 |
0.1670021276 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 10 |
Hb_005535_080 |
0.1674714383 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 11 |
Hb_001609_040 |
0.1677831603 |
- |
- |
hypothetical protein POPTR_0017s05650g [Populus trichocarpa] |
| 12 |
Hb_001268_340 |
0.1698998644 |
- |
- |
Actin, putative [Ricinus communis] |
| 13 |
Hb_000300_090 |
0.170995264 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_008406_180 |
0.1721190637 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 15 |
Hb_000193_300 |
0.1727557789 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 16 |
Hb_000008_190 |
0.1754371407 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_024074_020 |
0.1758203162 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 18 |
Hb_000889_100 |
0.1758248448 |
- |
- |
PREDICTED: syntaxin-132-like isoform X2 [Sesamum indicum] |
| 19 |
Hb_004126_040 |
0.1760023216 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_019181_030 |
0.1766654625 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |