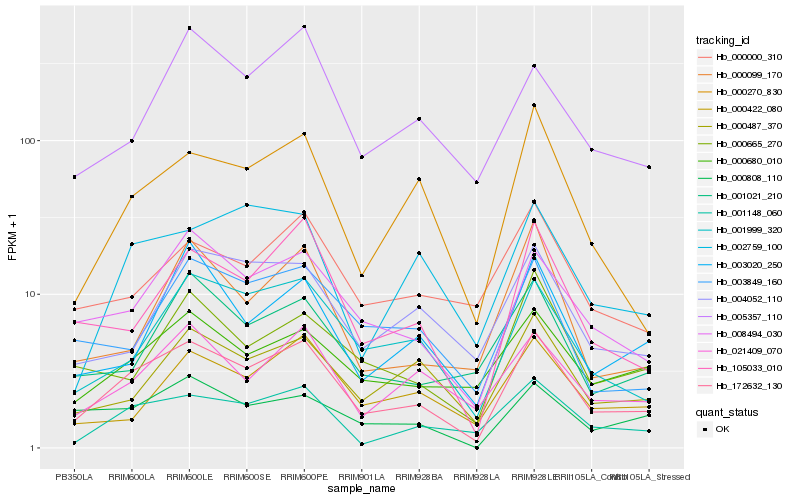
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172632_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_22584 [Jatropha curcas] |
| 2 |
Hb_008494_030 |
0.1294528724 |
- |
- |
catalytic, putative [Ricinus communis] |
| 3 |
Hb_001148_060 |
0.1379003178 |
- |
- |
hypothetical protein JCGZ_00278 [Jatropha curcas] |
| 4 |
Hb_001999_320 |
0.1400102426 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 5 |
Hb_105033_010 |
0.1438002139 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_021409_070 |
0.1513605234 |
- |
- |
PREDICTED: transcription factor bHLH157 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000270_830 |
0.1525693267 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_000487_370 |
0.1530201652 |
- |
- |
- |
| 9 |
Hb_000680_010 |
0.1561674644 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Populus euphratica] |
| 10 |
Hb_000422_080 |
0.1632004701 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 11 |
Hb_003849_160 |
0.1643143022 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 12 |
Hb_000665_270 |
0.1659492669 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 13 |
Hb_000808_110 |
0.1663863746 |
- |
- |
PREDICTED: uncharacterized protein LOC105647448 isoform X4 [Jatropha curcas] |
| 14 |
Hb_001021_210 |
0.1675894953 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005357_110 |
0.1682349173 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 16 |
Hb_003020_250 |
0.1687642925 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 17 |
Hb_000000_310 |
0.169208528 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 18 |
Hb_004052_110 |
0.1693100672 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000099_170 |
0.1701744648 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 20 |
Hb_002759_100 |
0.1705847685 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |