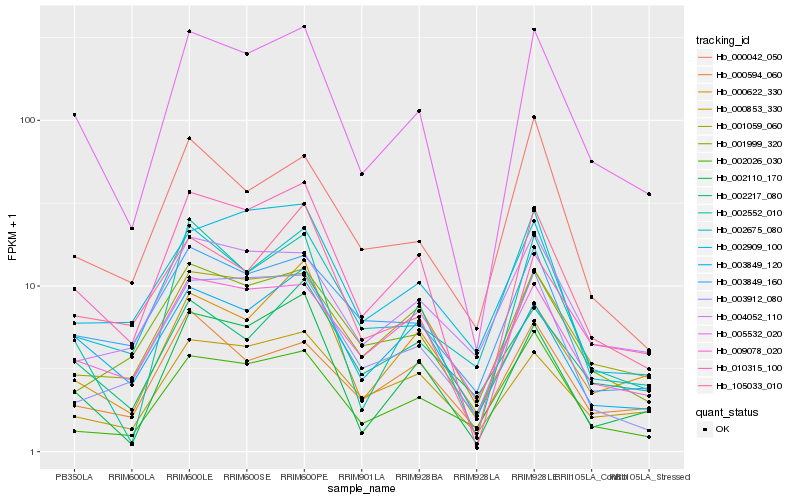
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005532_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100262861 [Vitis vinifera] |
| 2 |
Hb_002552_010 |
0.1208698338 |
transcription factor |
TF Family: DBP |
protein phosphatase 2c, putative [Ricinus communis] |
| 3 |
Hb_002110_170 |
0.1215681311 |
- |
- |
hypothetical protein CICLE_v10002406mg [Citrus clementina] |
| 4 |
Hb_105033_010 |
0.1281669586 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001059_060 |
0.1354351972 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 6 |
Hb_003849_120 |
0.1370750433 |
- |
- |
PREDICTED: uncharacterized protein LOC100264440 [Vitis vinifera] |
| 7 |
Hb_009078_020 |
0.1409673293 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 8 |
Hb_000853_330 |
0.1410418652 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 9 |
Hb_003849_160 |
0.1435456354 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 10 |
Hb_001999_320 |
0.1469283464 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 11 |
Hb_002675_080 |
0.1470455385 |
- |
- |
PREDICTED: uncharacterized protein LOC105634950 [Jatropha curcas] |
| 12 |
Hb_002026_030 |
0.1471450438 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 13 |
Hb_000622_330 |
0.1481383916 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 14 |
Hb_000042_050 |
0.1506072802 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000594_060 |
0.1518082438 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 16 |
Hb_010315_100 |
0.1527202926 |
- |
- |
PREDICTED: uncharacterized protein LOC104420162 [Eucalyptus grandis] |
| 17 |
Hb_002217_080 |
0.1529952904 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 16 [Jatropha curcas] |
| 18 |
Hb_003912_080 |
0.1566491488 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 19 |
Hb_002909_100 |
0.1579285199 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 20 |
Hb_004052_110 |
0.1580680986 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |