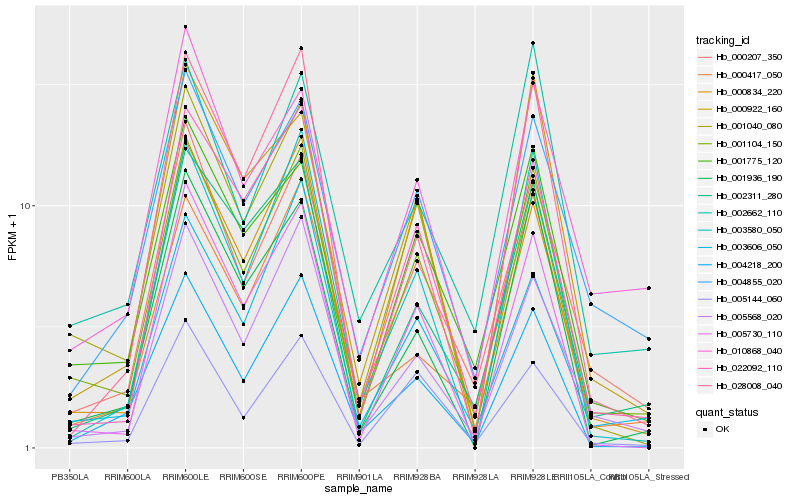
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001104_150 |
0.0 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 2 |
Hb_000207_350 |
0.083753189 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 3 |
Hb_001775_120 |
0.0925235892 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 4 |
Hb_005568_020 |
0.0997748848 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005730_110 |
0.1054588424 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 6 |
Hb_022092_110 |
0.1063615223 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 7 |
Hb_004218_200 |
0.1069849572 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 8 |
Hb_005144_060 |
0.1085615809 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 9 |
Hb_004855_020 |
0.1109704368 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 10 |
Hb_002311_280 |
0.1122930725 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 11 |
Hb_001936_190 |
0.1161606663 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 12 |
Hb_003580_050 |
0.1162169974 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 13 |
Hb_000922_160 |
0.1193701745 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 14 |
Hb_001040_080 |
0.1212557529 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 15 |
Hb_000417_050 |
0.1225540599 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 16 |
Hb_002662_110 |
0.1228435085 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 17 |
Hb_000834_220 |
0.1230650306 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 18 |
Hb_028008_040 |
0.1234733775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003606_050 |
0.1239472322 |
- |
- |
PREDICTED: beta-galactosidase 10 [Jatropha curcas] |
| 20 |
Hb_010868_040 |
0.1239782084 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |