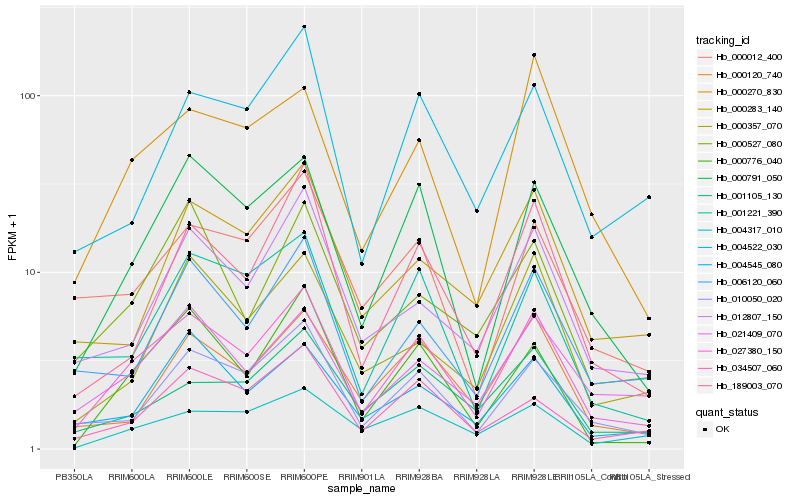
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027380_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000791_050 |
0.113140573 |
- |
- |
PREDICTED: uncharacterized protein LOC105636760 [Jatropha curcas] |
| 3 |
Hb_010050_020 |
0.1194356387 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 4 |
Hb_000776_040 |
0.1313732841 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X3 [Jatropha curcas] |
| 5 |
Hb_021409_070 |
0.1378657491 |
- |
- |
PREDICTED: transcription factor bHLH157 isoform X1 [Jatropha curcas] |
| 6 |
Hb_189003_070 |
0.1403337233 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 7 |
Hb_012807_150 |
0.141416869 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 8 |
Hb_006120_060 |
0.1417962763 |
- |
- |
O-acetyltransferase family protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_001221_390 |
0.1422321016 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 10 |
Hb_000120_740 |
0.1430663757 |
desease resistance |
Gene Name: VARLMGL |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000270_830 |
0.143257915 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_034507_060 |
0.1434633523 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic-like [Citrus sinensis] |
| 13 |
Hb_004545_080 |
0.1440780352 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_000012_400 |
0.1447054466 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 15 |
Hb_001105_130 |
0.1483699607 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 16 |
Hb_004522_030 |
0.1503349693 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 17 |
Hb_000527_080 |
0.1533395738 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 18 |
Hb_004317_010 |
0.1543704053 |
- |
- |
hypothetical protein CICLE_v10011038mg [Citrus clementina] |
| 19 |
Hb_000357_070 |
0.1553139804 |
- |
- |
- |
| 20 |
Hb_000283_140 |
0.1559466837 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |