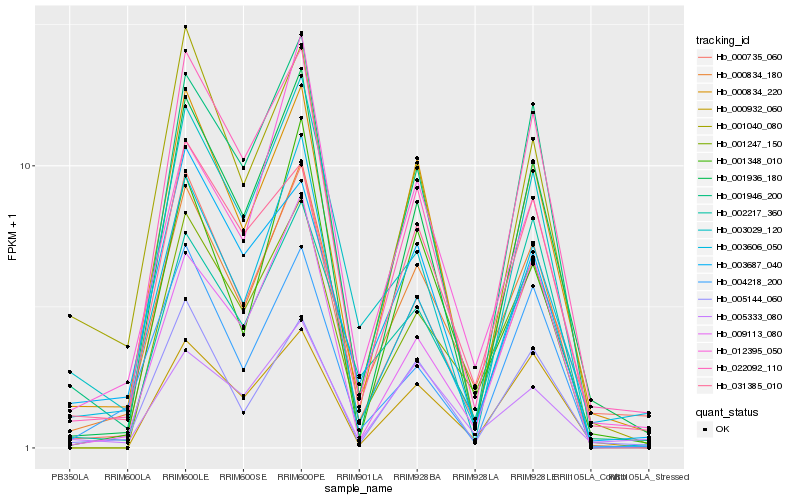
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000834_180 |
0.0 |
- |
- |
PREDICTED: kinesin-13A-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001247_150 |
0.1098551765 |
- |
- |
hypothetical protein JCGZ_23179 [Jatropha curcas] |
| 3 |
Hb_000834_220 |
0.1174147748 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 4 |
Hb_031385_010 |
0.1255197018 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 5 |
Hb_001946_200 |
0.1263999861 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 6 |
Hb_009113_080 |
0.1325731143 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 7 |
Hb_002217_360 |
0.1333016335 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003606_050 |
0.1358115661 |
- |
- |
PREDICTED: beta-galactosidase 10 [Jatropha curcas] |
| 9 |
Hb_022092_110 |
0.136121598 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 10 |
Hb_005144_060 |
0.1377104247 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 11 |
Hb_012395_050 |
0.1381656885 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 12 |
Hb_000735_060 |
0.1382248011 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 13 |
Hb_003687_040 |
0.1388195279 |
- |
- |
PREDICTED: uncharacterized protein LOC105632153 [Jatropha curcas] |
| 14 |
Hb_004218_200 |
0.1394193732 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 15 |
Hb_003029_120 |
0.1394253746 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 16 |
Hb_001040_080 |
0.1413614476 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 17 |
Hb_000932_060 |
0.141681236 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001936_180 |
0.1421567975 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 19 |
Hb_001348_010 |
0.1424741315 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 20 |
Hb_005333_080 |
0.1426653135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |