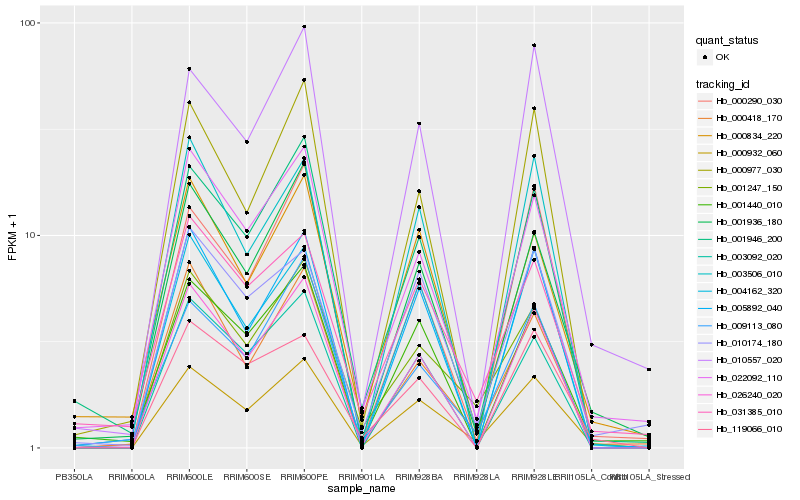
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000932_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001247_150 |
0.1034268576 |
- |
- |
hypothetical protein JCGZ_23179 [Jatropha curcas] |
| 3 |
Hb_001936_180 |
0.1062640356 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 4 |
Hb_022092_110 |
0.1121123893 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 5 |
Hb_010557_020 |
0.1126000089 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 6 |
Hb_026240_020 |
0.1153189723 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 7 |
Hb_001946_200 |
0.1162171858 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 8 |
Hb_004162_320 |
0.1191101612 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000418_170 |
0.119541786 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 10 |
Hb_010174_180 |
0.12050839 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_001440_010 |
0.1213350368 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 12 |
Hb_005892_040 |
0.1232692031 |
- |
- |
hypothetical protein JCGZ_02368 [Jatropha curcas] |
| 13 |
Hb_119066_010 |
0.1257457411 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 14 |
Hb_000290_030 |
0.1263128555 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 15 |
Hb_000834_220 |
0.1267274871 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 16 |
Hb_000977_030 |
0.1271610699 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_009113_080 |
0.1289034994 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 18 |
Hb_003506_010 |
0.1289493497 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 19 |
Hb_031385_010 |
0.1298867718 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 20 |
Hb_003092_020 |
0.1321263962 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |