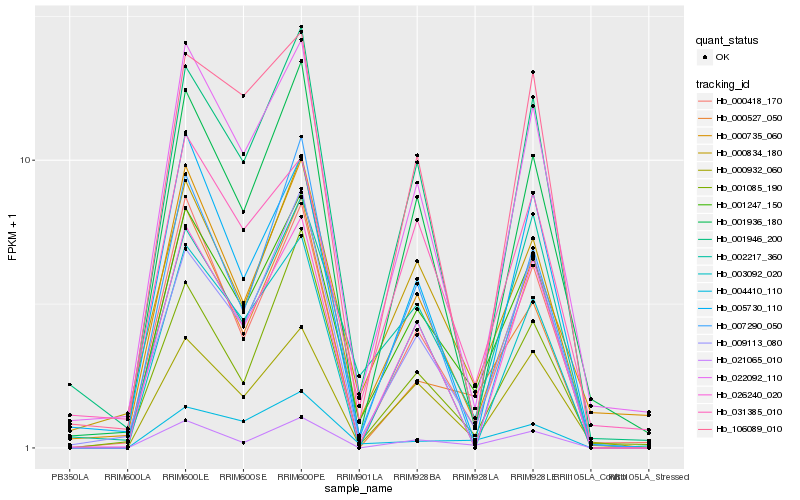
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001247_150 |
0.0 |
- |
- |
hypothetical protein JCGZ_23179 [Jatropha curcas] |
| 2 |
Hb_000932_060 |
0.1034268576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_021065_010 |
0.1071230149 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like isoform X2 [Gossypium raimondii] |
| 4 |
Hb_000834_180 |
0.1098551765 |
- |
- |
PREDICTED: kinesin-13A-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000418_170 |
0.1179871466 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 6 |
Hb_009113_080 |
0.1202369886 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 7 |
Hb_022092_110 |
0.1252409337 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 8 |
Hb_001946_200 |
0.1286318648 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 9 |
Hb_026240_020 |
0.135007558 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 10 |
Hb_001936_180 |
0.1390157156 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 11 |
Hb_002217_360 |
0.1390801147 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001085_190 |
0.1425879324 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 13 |
Hb_031385_010 |
0.1426423411 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 14 |
Hb_000527_050 |
0.1429215755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004410_110 |
0.1441229023 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_007290_050 |
0.1452898102 |
- |
- |
PREDICTED: uncharacterized protein LOC105629472 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003092_020 |
0.1470008081 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |
| 18 |
Hb_000735_060 |
0.1497636497 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 19 |
Hb_005730_110 |
0.1501099895 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 20 |
Hb_106089_010 |
0.1509623002 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |