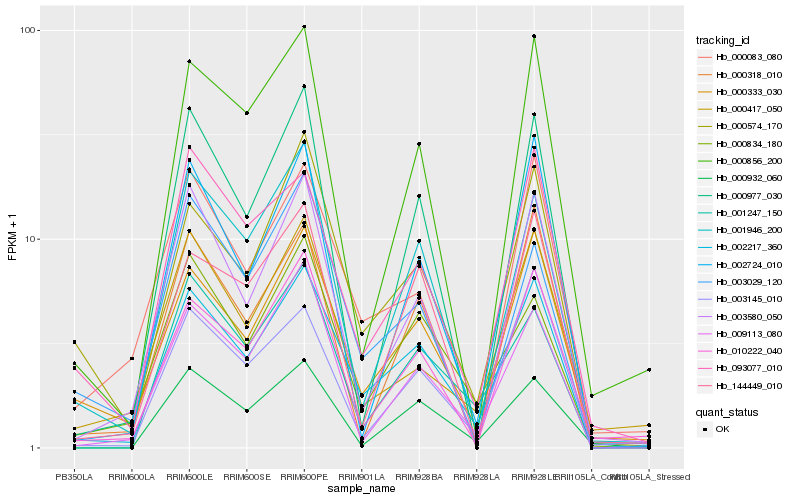
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_360 |
0.0 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000333_030 |
0.107347798 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000574_170 |
0.1218409801 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump-like [Jatropha curcas] |
| 4 |
Hb_010222_040 |
0.1278897939 |
- |
- |
PREDICTED: kinesin-2 [Jatropha curcas] |
| 5 |
Hb_000834_180 |
0.1333016335 |
- |
- |
PREDICTED: kinesin-13A-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001946_200 |
0.1348711733 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_093077_010 |
0.1372165872 |
transcription factor |
TF Family: G2-like |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_009113_080 |
0.1387031344 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 9 |
Hb_001247_150 |
0.1390801147 |
- |
- |
hypothetical protein JCGZ_23179 [Jatropha curcas] |
| 10 |
Hb_000083_080 |
0.141262268 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 11 |
Hb_003580_050 |
0.1418983806 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 12 |
Hb_000856_200 |
0.1446139726 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 13 |
Hb_144449_010 |
0.1453547678 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 14 |
Hb_000932_060 |
0.1492589507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000417_050 |
0.1495121746 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 16 |
Hb_003029_120 |
0.1498824611 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 17 |
Hb_000977_030 |
0.1519466269 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_002724_010 |
0.1530462637 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 19 |
Hb_003145_010 |
0.1549725518 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 20 |
Hb_000318_010 |
0.1562771774 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |