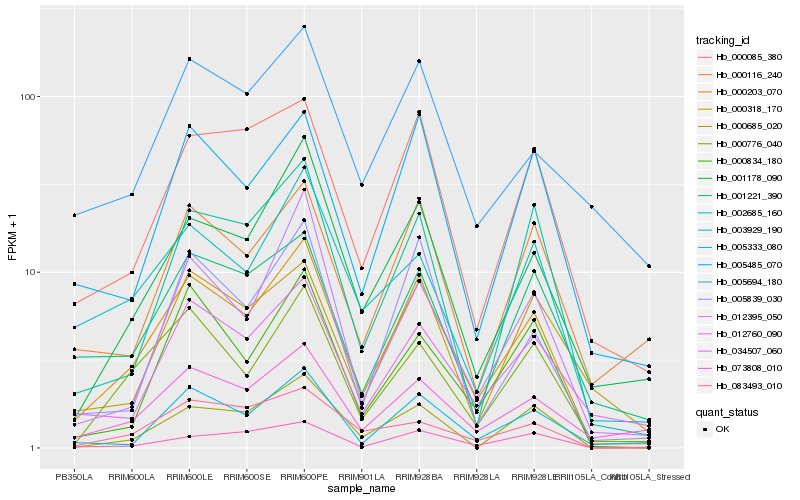
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000203_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649674 [Jatropha curcas] |
| 2 |
Hb_000776_040 |
0.1179050523 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X3 [Jatropha curcas] |
| 3 |
Hb_001178_090 |
0.1309762507 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 4 |
Hb_034507_060 |
0.1325565997 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic-like [Citrus sinensis] |
| 5 |
Hb_005333_080 |
0.1351468423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000085_380 |
0.1353754141 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_012760_090 |
0.1359441918 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 8 |
Hb_005485_070 |
0.1402780801 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_002685_160 |
0.1418088894 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 10 |
Hb_003929_190 |
0.1434937419 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 11 |
Hb_000834_180 |
0.1436778985 |
- |
- |
PREDICTED: kinesin-13A-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001221_390 |
0.1445256915 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 13 |
Hb_000685_020 |
0.146721011 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 4-like [Jatropha curcas] |
| 14 |
Hb_005839_030 |
0.1473721649 |
- |
- |
PREDICTED: potassium transporter 2 [Jatropha curcas] |
| 15 |
Hb_012395_050 |
0.1504832678 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 16 |
Hb_083493_010 |
0.1523826864 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 17 |
Hb_073808_010 |
0.1550197351 |
desease resistance |
Gene Name: DUF594 |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_000116_240 |
0.1570356561 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 19 |
Hb_000318_170 |
0.1574415794 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 20 |
Hb_005694_180 |
0.1592027199 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |