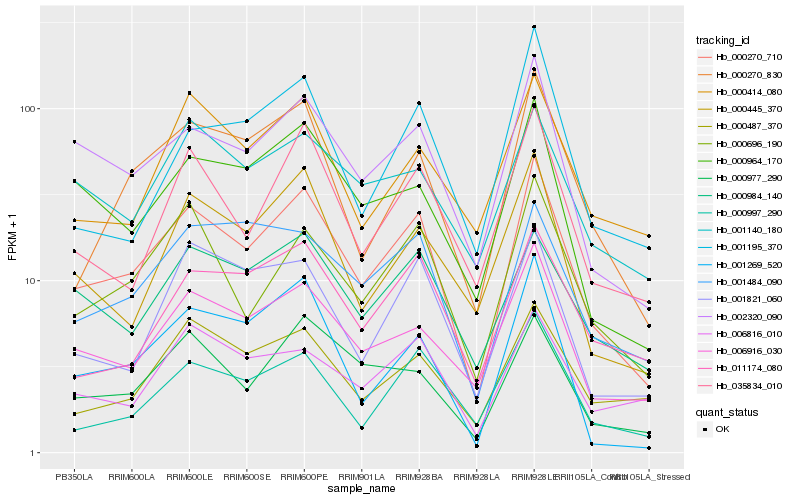
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_710 |
0.0 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002320_090 |
0.1057198066 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 3 |
Hb_000696_190 |
0.1179993545 |
- |
- |
PREDICTED: protein MID1-COMPLEMENTING ACTIVITY 1 [Jatropha curcas] |
| 4 |
Hb_006916_030 |
0.1188068458 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 5 |
Hb_000270_830 |
0.1252736884 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_035834_010 |
0.1281766714 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_000964_170 |
0.1300326516 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 8 |
Hb_000997_290 |
0.1313954328 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 9 |
Hb_000445_370 |
0.1326090054 |
- |
- |
PREDICTED: probable methyltransferase PMT24 [Jatropha curcas] |
| 10 |
Hb_000977_290 |
0.1334400692 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_001269_520 |
0.1372483366 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 12 |
Hb_001484_090 |
0.1415888739 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_001821_060 |
0.1424541746 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 14 |
Hb_000984_140 |
0.1436909799 |
- |
- |
PREDICTED: xylulose kinase [Jatropha curcas] |
| 15 |
Hb_000487_370 |
0.1448975239 |
- |
- |
- |
| 16 |
Hb_011174_080 |
0.145150089 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 17 |
Hb_000414_080 |
0.1457582467 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_006816_010 |
0.1485919352 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001140_180 |
0.1490431569 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 20 |
Hb_001195_370 |
0.1499990645 |
- |
- |
- |