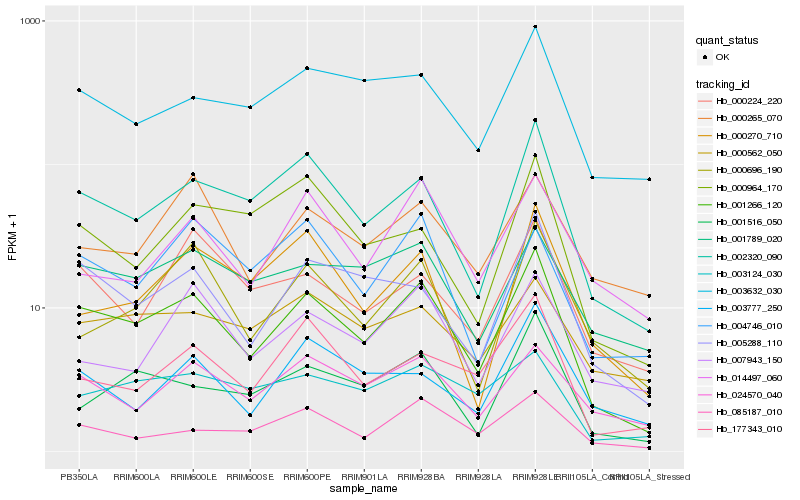
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001266_120 |
0.0 |
- |
- |
auxilin, putative [Ricinus communis] |
| 2 |
Hb_002320_090 |
0.1135545215 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 3 |
Hb_005288_110 |
0.1358580831 |
- |
- |
PREDICTED: uncharacterized protein LOC105647752 [Jatropha curcas] |
| 4 |
Hb_024570_040 |
0.153122606 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 5 |
Hb_003632_030 |
0.1552973883 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 6 |
Hb_085187_010 |
0.156195949 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 7 |
Hb_177343_010 |
0.156327162 |
- |
- |
PREDICTED: uncharacterized protein LOC105650963 [Jatropha curcas] |
| 8 |
Hb_001516_050 |
0.1578015173 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 9 |
Hb_001789_020 |
0.1584930071 |
- |
- |
PREDICTED: tripeptidyl-peptidase 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000696_190 |
0.158839393 |
- |
- |
PREDICTED: protein MID1-COMPLEMENTING ACTIVITY 1 [Jatropha curcas] |
| 11 |
Hb_003124_030 |
0.1630810057 |
- |
- |
At4g33625-like protein [Brassica napus] |
| 12 |
Hb_000224_220 |
0.1640166712 |
- |
- |
PREDICTED: protein disulfide isomerase-like 2-3 [Jatropha curcas] |
| 13 |
Hb_000270_710 |
0.1649282628 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000562_050 |
0.1692899329 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 15 |
Hb_004746_010 |
0.1705976518 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 16 |
Hb_007943_150 |
0.1706496595 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_000265_070 |
0.1717531664 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 18 |
Hb_000964_170 |
0.1733365618 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 19 |
Hb_014497_060 |
0.174541098 |
- |
- |
phosphofructokinase, putative [Ricinus communis] |
| 20 |
Hb_003777_250 |
0.1746587834 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |