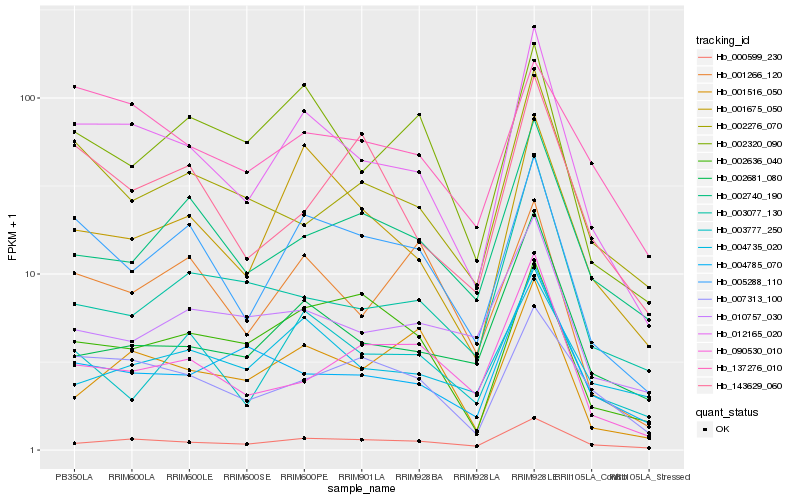
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012165_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 2 |
Hb_005288_110 |
0.1352947517 |
- |
- |
PREDICTED: uncharacterized protein LOC105647752 [Jatropha curcas] |
| 3 |
Hb_000599_230 |
0.1400506695 |
- |
- |
- |
| 4 |
Hb_001675_050 |
0.1455702353 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 5 |
Hb_007313_100 |
0.1548203065 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 6 |
Hb_002681_080 |
0.1604578142 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002276_070 |
0.1670220233 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 8 |
Hb_002320_090 |
0.1713010414 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 9 |
Hb_004785_070 |
0.1727895153 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_137276_010 |
0.1780614322 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 11 |
Hb_003777_250 |
0.1791729398 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 12 |
Hb_002740_190 |
0.1821278405 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_090530_010 |
0.1826356384 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 14 |
Hb_143629_060 |
0.1843152703 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001516_050 |
0.1856389659 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 16 |
Hb_003077_130 |
0.1862000875 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 17 |
Hb_010757_030 |
0.1865676666 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_004735_020 |
0.1883471642 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 19 |
Hb_001266_120 |
0.1894150129 |
- |
- |
auxilin, putative [Ricinus communis] |
| 20 |
Hb_002636_040 |
0.1903543477 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |