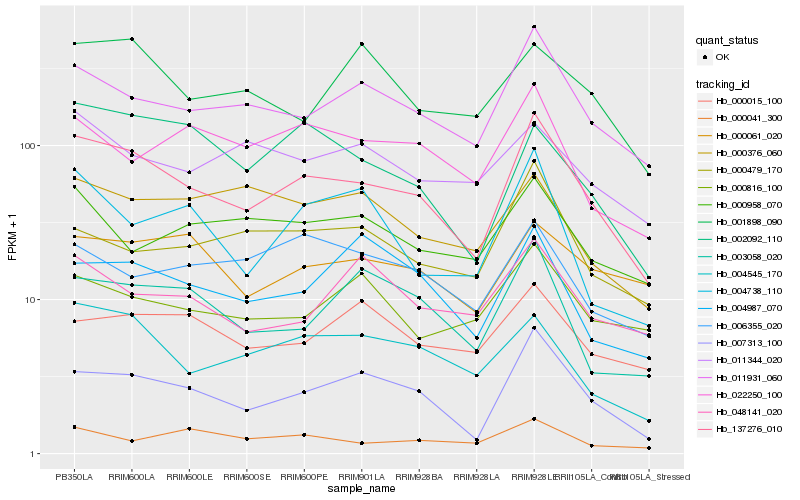
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_137276_010 |
0.0 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 2 |
Hb_007313_100 |
0.1017257984 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 3 |
Hb_011931_060 |
0.1109868688 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 4 |
Hb_004545_170 |
0.1377379963 |
- |
- |
PREDICTED: autophagy-related protein 18h [Jatropha curcas] |
| 5 |
Hb_003058_020 |
0.1515539191 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_022250_100 |
0.1521222961 |
- |
- |
calnexin, putative [Ricinus communis] |
| 7 |
Hb_002092_110 |
0.152748743 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 8 |
Hb_000061_020 |
0.1552381384 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 9 |
Hb_000479_170 |
0.1556451638 |
- |
- |
chaperone clpb, putative [Ricinus communis] |
| 10 |
Hb_004738_110 |
0.1583999488 |
- |
- |
phosphoenolpyruvate carboxylase [Manihot esculenta] |
| 11 |
Hb_004987_070 |
0.1585347922 |
- |
- |
PREDICTED: heat shock 70 kDa protein 17-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000958_070 |
0.1592277637 |
- |
- |
hypothetical protein PRUPE_ppa007681mg [Prunus persica] |
| 13 |
Hb_000816_100 |
0.159701523 |
- |
- |
PREDICTED: APO protein 1, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_006355_020 |
0.1612949401 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 15 |
Hb_011344_020 |
0.1619487871 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 16 |
Hb_000041_300 |
0.1627855021 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized ATP-dependent helicase C23E6.02 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000015_100 |
0.1628858078 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_048141_020 |
0.1633000062 |
- |
- |
tetratricopeptide repeat-like superfamily protein, partial [Manihot esculenta] |
| 19 |
Hb_001898_090 |
0.1633592271 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000376_060 |
0.1653216952 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |