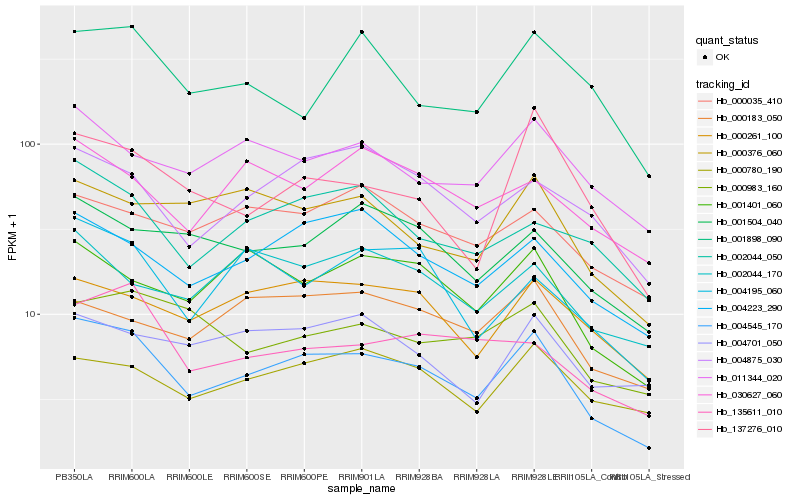
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004545_170 |
0.0 |
- |
- |
PREDICTED: autophagy-related protein 18h [Jatropha curcas] |
| 2 |
Hb_004223_290 |
0.1244805279 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 3 |
Hb_002044_050 |
0.1315523285 |
- |
- |
- |
| 4 |
Hb_011344_020 |
0.1339080219 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 5 |
Hb_000261_100 |
0.1344019563 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001401_060 |
0.134545231 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 7 |
Hb_004875_030 |
0.1345988643 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 8 |
Hb_001504_040 |
0.1362326948 |
- |
- |
PREDICTED: factor of DNA methylation 1-like [Jatropha curcas] |
| 9 |
Hb_000780_190 |
0.1374378374 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 10 |
Hb_137276_010 |
0.1377379963 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 11 |
Hb_004195_060 |
0.1396556965 |
- |
- |
PREDICTED: uncharacterized protein LOC105803780 isoform X1 [Gossypium raimondii] |
| 12 |
Hb_000376_060 |
0.1413688876 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_135611_010 |
0.1415170267 |
- |
- |
PREDICTED: uncharacterized protein LOC105142085 [Populus euphratica] |
| 14 |
Hb_004701_050 |
0.145676729 |
- |
- |
alpha1 family protein [Populus trichocarpa] |
| 15 |
Hb_000035_410 |
0.1463648805 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 16 |
Hb_000983_160 |
0.1468916324 |
- |
- |
hypothetical protein B456_009G199800 [Gossypium raimondii] |
| 17 |
Hb_030627_060 |
0.1474704107 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 18 |
Hb_002044_170 |
0.1477599564 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 19 |
Hb_001898_090 |
0.147886871 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000183_050 |
0.1495153041 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |