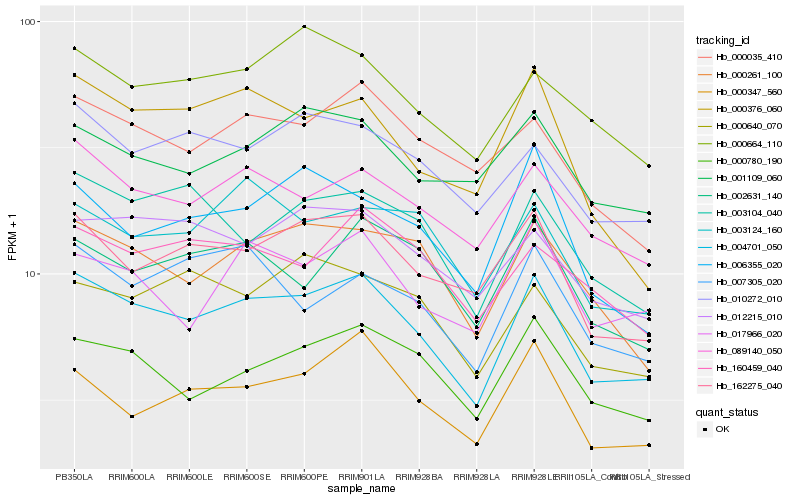
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004701_050 |
0.0 |
- |
- |
alpha1 family protein [Populus trichocarpa] |
| 2 |
Hb_089140_050 |
0.0785329594 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 3 |
Hb_162275_040 |
0.0802697955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000261_100 |
0.082686518 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000780_190 |
0.0837260773 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 6 |
Hb_010272_010 |
0.0873413609 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 7 |
Hb_000035_410 |
0.0880807749 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 8 |
Hb_003104_040 |
0.0884227249 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 9 |
Hb_000347_560 |
0.0892647425 |
- |
- |
- |
| 10 |
Hb_012215_010 |
0.0896372229 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 11 |
Hb_000376_060 |
0.091566407 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000664_110 |
0.0920976529 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 13 |
Hb_002631_140 |
0.0942014211 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 12, chloroplastic [Jatropha curcas] |
| 14 |
Hb_017966_020 |
0.0950526666 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03560, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000640_070 |
0.0955741458 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 16 |
Hb_007305_020 |
0.0957806442 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 17 |
Hb_003124_160 |
0.0958362576 |
- |
- |
dynamin, putative [Ricinus communis] |
| 18 |
Hb_006355_020 |
0.0967185088 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 19 |
Hb_160459_040 |
0.0968041333 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 20 |
Hb_001109_060 |
0.0969225976 |
- |
- |
amino acid transporter, putative [Ricinus communis] |