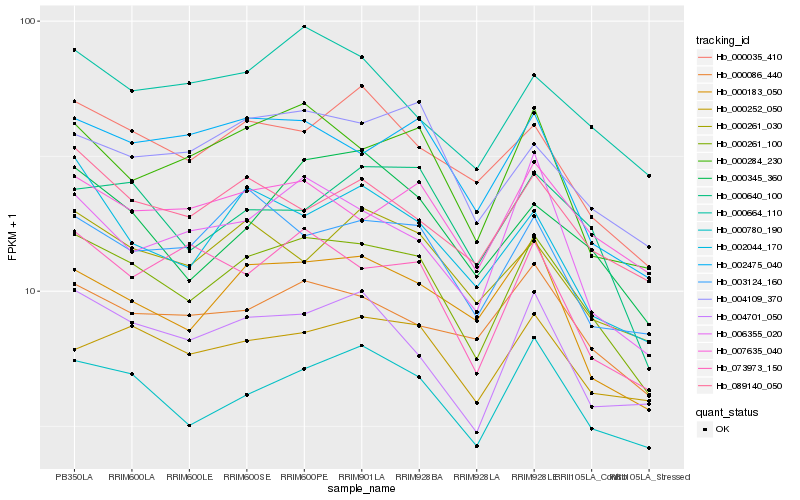
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_100 |
0.0 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000780_190 |
0.0695562099 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 3 |
Hb_000284_230 |
0.0808710486 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 4 |
Hb_000035_410 |
0.0819977642 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 5 |
Hb_004701_050 |
0.082686518 |
- |
- |
alpha1 family protein [Populus trichocarpa] |
| 6 |
Hb_000183_050 |
0.0831726221 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 7 |
Hb_004109_370 |
0.0834240479 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 8 |
Hb_000640_100 |
0.0840508791 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000086_440 |
0.0850024001 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 10 |
Hb_002475_040 |
0.0854244321 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 11 |
Hb_007635_040 |
0.087240747 |
- |
- |
PREDICTED: WAT1-related protein At3g45870 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000345_360 |
0.0873289036 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |
| 13 |
Hb_000252_050 |
0.0885012737 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 14 |
Hb_089140_050 |
0.0885722543 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 15 |
Hb_003124_160 |
0.0892754178 |
- |
- |
dynamin, putative [Ricinus communis] |
| 16 |
Hb_000261_030 |
0.089406329 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 17 |
Hb_000664_110 |
0.0894760807 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 18 |
Hb_073973_150 |
0.091252967 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002044_170 |
0.0918060035 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 20 |
Hb_006355_020 |
0.0918275109 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |