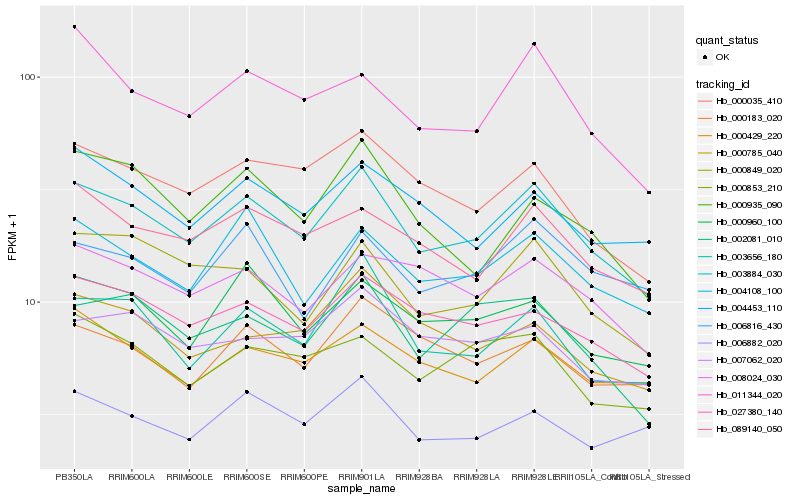
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003884_030 |
0.0 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 2 |
Hb_027380_140 |
0.0729229715 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000035_410 |
0.0747547759 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 4 |
Hb_089140_050 |
0.0757074552 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 5 |
Hb_004108_100 |
0.0762652429 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X2 [Jatropha curcas] |
| 6 |
Hb_003656_180 |
0.0766677224 |
- |
- |
PREDICTED: uncharacterized protein LOC105637927 [Jatropha curcas] |
| 7 |
Hb_000429_220 |
0.0795195169 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000960_100 |
0.0797169348 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 9 |
Hb_002081_010 |
0.0816922205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008024_030 |
0.0817432892 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 11 |
Hb_000849_020 |
0.0832187581 |
- |
- |
PREDICTED: uncharacterized protein LOC105644228 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004453_110 |
0.0838730576 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 13 |
Hb_011344_020 |
0.0844203306 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 14 |
Hb_000853_210 |
0.0849560196 |
- |
- |
PREDICTED: uncharacterized protein LOC105642575 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007062_020 |
0.08645115 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 16 |
Hb_000183_020 |
0.0881138036 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006882_020 |
0.0881352221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000935_090 |
0.0904244517 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 19 |
Hb_000785_040 |
0.0905165105 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 20 |
Hb_006816_430 |
0.0911999979 |
- |
- |
PREDICTED: topless-related protein 3 isoform X1 [Jatropha curcas] |